Report
Scientific Title
Exploring the Role of Inflammatory Indices in Predicting the Risk of Non-Alcoholic Fatty Liver Disease or Cirrhosis Among Healthy Adults Using UK Biobank Data
Group Members
Zicheng Wang (zw3088), Xiaohui Kang (xk2163), Mengyuan Chen (mc5698), Shuai yuan (sy3270), Dang Lin (dl3757)
Motivation
Fatty liver disease (FLD) is one of the most prevalent chronic diseases and includes non-alcoholic fatty liver disease (NAFLD), metabolic-associated fatty liver disease (MAFLD), and metabolic-associated steatohepatitis-related liver disease (MASLD). Among these, NAFLD is the most common form, and its prevalence is rising rapidly. This condition can progress to severe complications, such as cirrhosis, liver failure, or hepatocellular carcinoma. Systemic inflammation and immune dysregulation are closely associated with FLD, underscoring the need to identify reliable, non-invasive biomarkers. Such biomarkers are critical for predicting disease progression, understanding underlying mechanisms, enabling early detection, guiding interventions, and ultimately improving clinical outcomes.
Immune-related indices like the Lymphocyte-to-Monocyte Ratio (LMR), Systemic Immune-Inflammation Index (SII), Naples Prognostic Score (NPS), and Neutrophil-Percentage-to-Albumin Ratio (NPAR) have gained attention for their potential utility in assessing inflammation and immune responses. These indices are derived from routine blood tests, making them accessible and cost-effective. LMR, for instance, measures the balance between lymphocytes, which are linked to immune surveillance and anti-inflammatory responses, and monocytes, which are associated with inflammation and tissue damage. SII integrates neutrophil, platelet, and lymphocyte counts to provide a comprehensive view of immune activity and thrombosis risk. NPS combines multiple prognostic factors, including inflammatory markers, albumin levels, and platelet counts, offering a broader perspective on patient health. NPAR evaluates the relationship between neutrophil percentages and albumin levels, connecting immune function with nutritional status.
The UK Biobank (UKB) dataset is well-suited for studying the relationships between these immune markers and chronic disease outcomes. With its extensive biomarker, demographic, and longitudinal health data on a representative UK population, the UKB provides an opportunity for detailed cross-sectional and longitudinal analyses. These analyses could help clarify how inflammation and immune stress contribute to chronic disease progression, ultimately advancing both clinical and public health approaches.
Initial Questions
Our initial questions were structured to explore potential connections between variables and their relationships to specific indices and chronic diseases:
What are the initial associations between key variables such as age, sex, alcohol consumption, blood pressure, income, diet quality, and sleep duration?
Are there significant relationships between participants’ characteristics (e.g., age, sex, lifestyle factors) and the indices we aim to study (e.g., systemic immune-inflammation index or metabolic markers)?
Can these indices serve as predictors for specific chronic diseases, such as non-alcoholic fatty liver disease (NAFLD), and how do they interact with other variables in forecasting risk?
Data Summary
UKB Dataset
We utilized the UK Biobank (UKB) dataset, a large-scale biomedical database and research resource containing in-depth genetic and health information from over half a million UK participants. Following a formal data application process, we obtained permission to access and utilize the UKB dataset for our research. UK Biobank Dataset The dataset includes comprehensive health data collected through physical measurements, biological samples, and self-reported questionnaires, complemented by linked electronic health records. The data was initially collected between 2006 and 2010, with ongoing updates to include new information on health outcomes and laboratory tests as well as long-term follow-ups.
After the data cleaning steps, the cleaned dataset includes 413,146 observations and 113 variables. Although we will not use all the variables in this dataset, we will select multiple key variables to investigate our research questions:
expo_npar_cat: Neutrophil-to-albumin ratio categorized
into quartiles (Q1 = lowest, Q4 = highest).
age: Participants’ age, categorized into 10-year age
groups (e.g., 30–39, 40–49, etc.).
sex: Male or female.
income: Seven income brackets, ranging from 1 (lowest)
to 7 (highest).
townsend: Townsend deprivation index, a measure of
socioeconomic status, where higher values indicate greater deprivation.
total_met: Total metabolic equivalent of task score,
derived from physical activity data and grouped into categories.
diet_quality: Diet quality score, ranging from 0
(poorest) to 7 (highest), based on adherence to DASHguideline.
sleep_hour: Nightly sleep duration, measured in hours.
smoke_cat: Smoking status, categorized as current
smoker, past smoker, or non-smoker.
total_alcohol: Total alcohol consumption (ml/week),
grouped into categories (non-drinker, low, moderate, high, very high).
non_hdl: Non-HDL cholesterol level, a continuous
variable indicating cardiovascular risk. (mg/dl)
tg: Triglyceride levels, a continuous measure of blood
lipid levels. (mg/dl)
bp_cat: Blood pressure, categorized into intervals
(e.g., BP=20, 40, 60, etc.).
Exposure Variables
Lymphocyte-to-Monocyte Ratio (LMR): \[\text{LMR} = \frac{\text{Absolute Lymphocyte Count}}{\text{Absolute Monocyte Count}}\]
Systemic Immune-Inflammation Index (SII): \[\text{SII} = \frac{\text{Neutrophil Count} \times \text{Platelet Count}}{\text{Lymphocyte Count}}\]
Naples Prognostic Score (NPS): This score is based on a combination of factors, typically including:
- Albumin level (<4 g/dL = 1 point).
- Neutrophil-to-Lymphocyte Ratio (NLR) (>3 = 1 point).
- Lymphocyte-to-Monocyte Ratio (LMR) (<4.44 = 1 point).
- Platelet-to-Lymphocyte Ratio (PLR) (>150 = 1 point).
The final NPS is the sum of these points (range: 0–4).
Neutrophil-Percentage-to-Albumin Ratio (NPAR): \[\text{NPAR} = \frac{\text{Neutrophil Percentage}}{\text{Albumin Level (g/dL)}}\]
Note: The variables were categorized into tertiles based on the 25% and 75% percentiles after being ranked in ascending order, except for NPS, which was dichotomized based on whether it was ≥ 2.
Data Cleaning
Data Cleaning Part One
View Full Part One Data Cleaning Steps on GitHub
The pre-cleaning process involved several key steps to transform the
raw UKB dataset into a clean, analyzable format. Initially, variables
were renamed to improve clarity and usability. For instance,
n_21022_0_0 was renamed to age,
n_31_0_0 to sex, and n_21000_0_0
to ethnicity. New variables were derived from existing data
to enhance analysis potential. For example, physical activity metrics
such as MET_walk (n_22037_0_0),
MET_moderate (n_22038_0_0), and
MET_vigorous (n_22039_0_0) were combined to
create total_MET, which was further categorized into
activity levels using pa_cat. Invalid or missing data
values, such as -3 and -1, were replaced with
. (missing), while specific imputations were applied where
necessary, such as assigning 0.5 to dietary variables like
cooked_vegetable_0 when valid data was unavailable.
Variables were categorized for analytical ease, such as grouping
physical activity, sleep, and dietary patterns. Dietary components like
vegetables, fruits, fish, red meat, and grains were processed to create
binary indicators of “healthy” consumption (e.g.,
healthy_vegetable for ≥3 servings/day of vegetables and
healthy_red_meat for ≤1.5 servings/week of red meat). An
aggregate dietary quality score (diet_quality) was computed
by summing these indicators and further categorized into quality
levels.
For sleep patterns, multiple variables such as chronotype
(Chronotype_preference_0), insomnia
(Insomnia_0), snoring (snoring_0), and daytime
sleepiness (daytime_sleepiness_0) were processed to
calculate a composite sleep quality score (sleep_quality).
This score was then combined with categorized sleep duration
(sleep_cat) to refine the assessment of overall sleep
health.
Lifestyle factors like smoking and alcohol consumption were also
carefully processed. Smoking history variables
(smoke_current and smoke_past) were combined
to categorize individuals’ smoking behavior into smoke_cat.
Alcohol consumption was calculated using variables like
redwine_week, beer_week, and
spirits_week, and total intake was categorized into risk
levels.
Biomarkers such as lipid and glucose levels were processed to
generate new variables like non_hdl (non-HDL cholesterol
derived from tc and hdl) and
glu_cat (glucose levels categorized based on clinical
thresholds). Blood pressure variables (sbp and
dbp) were similarly used to classify individuals into
health risk categories.
The cleaned and processed dataset, containing all these refined and derived variables, was saved as a new csv to be used for further analysis. This comprehensive pre-cleaning ensured that the dataset was both reliable and tailored to support the research objectives.
Data Cleaning Part Two
View Full Part Two Data Cleaning Steps on GitHub
The subsequent data cleaning process was designed to ensure the
dataset’s quality and readiness for complex analyses, including
multi-state modeling. The process began with importing the csv file
exported in the previous steps and standardizing variable names using
janitor::clean_names() to improve readability and
usability. Key variables were then transformed and categorized. For
instance, sex was converted into a factor variable with 0
representing female and 1 representing male, while
ethnicity_cat was categorized as 0 for white and 1 for
others. Age was grouped into meaningful categories
(age_cat): <50, 50–59,
60–69, and ≥70. Socioeconomic variables, like
income, and lifestyle variables, such as smoking status
(smoke_cat), were also transformed into categorical
factors.
Participants with pre-existing conditions at baseline were excluded
to ensure accurate survival and progression analyses. This included
those with non-alcoholic fatty liver disease (NAFLD) or cirrhosis,
identified through the variables nafld_date and
cirrho_date. Outcome variables were created to classify
participants’ health status. For example, nafld_outcome and
cirrho_outcome assigned 0 for healthy participants and 1
for those who developed the disease post-baseline. Missing disease dates
were carefully imputed using other known dates, such as the death date
(death_date) or lost-to-follow-up date
(lost_date). For participants with no recorded outcomes, a
default date of 2022-10-31 was used to mark the end of
follow-up.
To calculate disease progression metrics, survival durations were
computed from the baseline to either disease diagnosis or the end of
follow-up. These metrics were stored as nafld_surv_duration
and cirrho_surv_duration. Blood test variables were also
renamed and cleaned, including lymphocytes (b_lympho),
monocytes (b_mono), platelets (b_plate), and
neutrophils (b_neutro). Observations with missing values in
these variables were dropped to ensure reliable downstream analyses.
New biomarkers were derived to assess participants’ health status and
inflammation levels. For example, the lymphocyte-to-monocyte ratio
(expo_lmr), systemic immune-inflammation index
(expo_sii), and neutrophil-to-albumin ratio
(expo_npar) were calculated. Critical thresholds were
applied to categorize these biomarkers into clinical risk groups, such
as low albumin levels (cri_album <4 g/dL) or high
neutrophil-to-lymphocyte ratios (cri_nlr >3). Biomarkers
were further categorized into quartiles using a custom function
(cut_1_23_4) to allow for stratified analyses.
The final dataset included essential variables, such as demographics
(sex, age), lifestyle factors
(smoke_cat, total_alcohol), biomarkers (e.g.,
tc for total cholesterol, hdl for high-density
lipoprotein, hba1c for blood glucose), and disease
outcomes. These variables were selected and exported as
data_prepared.dta. This ensured the dataset was robust,
consistent, and ready for both descriptive and inferential analyses,
particularly for modeling disease progression pathways.
Exploratory Data Analysis
To understand the data structure and for further EDA, we initially examined the associations between key demographic and lifestyle variables, such as age, sex, alcohol consumption, blood pressure, income, diet quality, and sleep duration. This step aimed to identify potential patterns and relationships between participants’ characteristics and overall health outcomes, laying the groundwork for subsequent analyses.
To facilitate the initial understanding of inflammatory indices and for further survival analysis, we explored the relationships between blood parameters (e.g., lymphocyte-to-monocyte ratio, systemic immune-inflammation index) and exposure variables with disease outcomes like NAFLD and cirrhosis. This step provided insights into the predictive value of these indices and their interactions with other variables, offering a foundation for examining disease progression in later stages of the project.
ukb_data <- read_dta("./external_data/data_prepared.dta")Age and Sex Distribution
figure_1 <- ukb_data %>%
mutate(age_group = cut(age, breaks = seq(10, 100, by = 10), right = FALSE,
labels = paste(seq(10, 90, by = 10), "-",
seq(19, 99, by = 10), sep = ""))) %>%
ggplot(aes(x = age_group, y = age, fill = as.factor(sex))) +
geom_boxplot(alpha = 0.9) +
theme_minimal() +
labs(x = 'Age Groups',
y = 'Age',
title = 'Age Distribution by Sex and Age Groups',
fill = 'Sex') +
scale_fill_manual(values = c("lightblue", "lightpink"),
labels = c("Female", "Male")) +
theme(legend.position = "bottom")
figure_1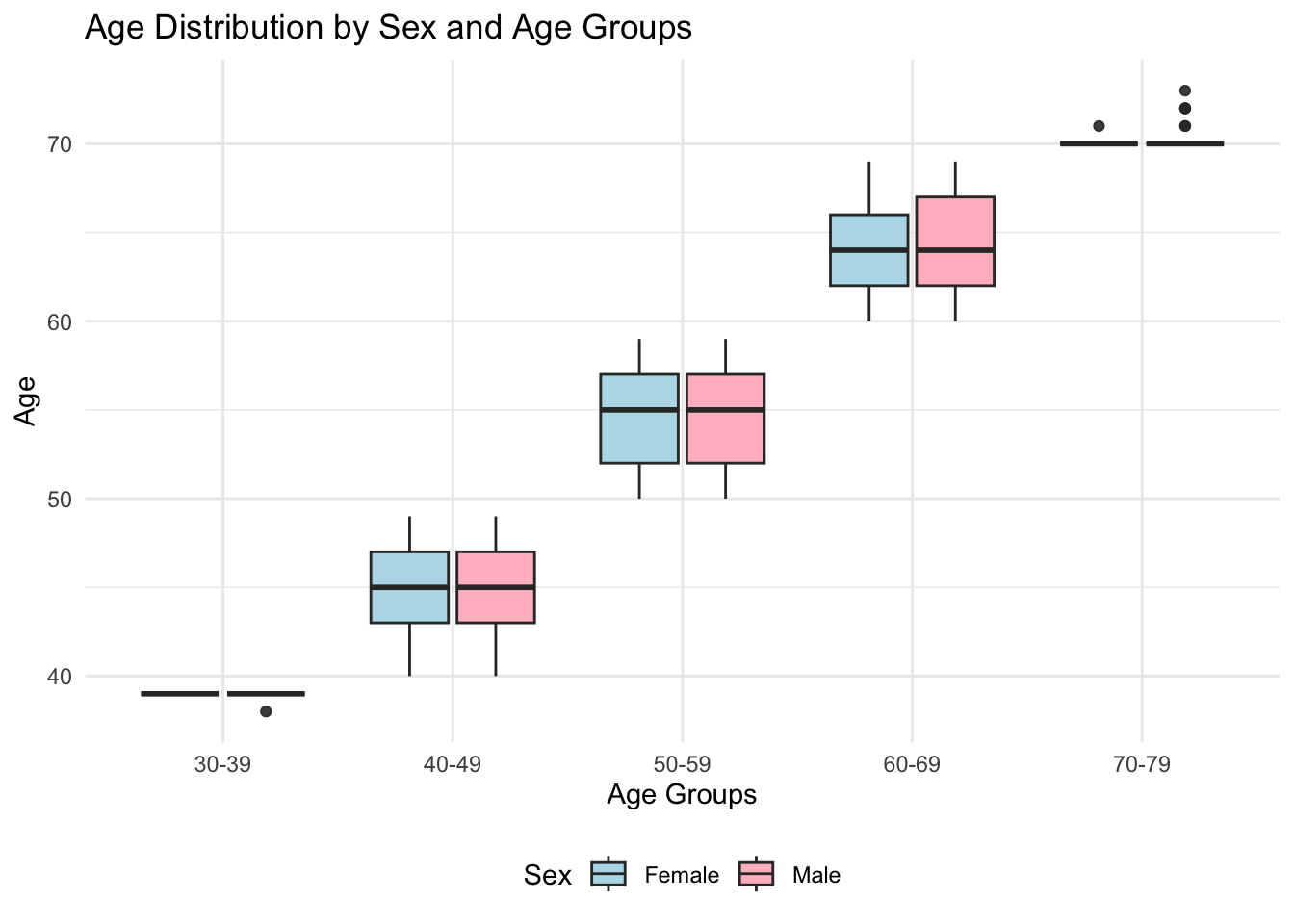
We examined the age distribution of male and female participants across 10-year age groups. In the 30-39 age group, the sample size was relatively small, with median ages and IQRs consistent across sexes. Similar trends were observed in the 40-49 and 50-59 groups. In the 60-69 group, females exhibited slightly higher upper quartiles than males, while the 70-79 group showed broader distributions for females, indicating more variation in ages.
Alcohol Consumption and Disease Incidence
# Data cleaning
disease_heat2 <- ukb_data %>%
filter(!is.na(sex), !is.na(total_alcohol), !is.na(nafld_outcome)) %>%
mutate(
alcohol_group = cut(total_alcohol,
breaks = c(-Inf, 0, 5, 10, 15, Inf),
labels = c("None", "Low", "Medium", "High", "Very High"),
include.lowest = TRUE)
) %>%
group_by(sex, alcohol_group) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
ukb_data <- ukb_data %>%
mutate(
sex = as_factor(sex),
nafld_outcome = as.numeric(as_factor(nafld_outcome)),
total_alcohol = as.numeric(total_alcohol),
income = as.numeric(income)
)
disease_heat2 <- ukb_data %>%
filter(!is.na(sex), !is.na(total_alcohol), !is.na(nafld_outcome)) %>%
mutate(
alcohol_group = cut(total_alcohol,
breaks = c(-Inf, 0, 5, 10, 15, Inf),
labels = c("None", "Low", "Medium", "High", "Very High"),
include.lowest = TRUE)
) %>%
group_by(sex, alcohol_group) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
# Create heatmap
figure_2 <- ggplot(disease_heat2, aes(x = sex, y = alcohol_group, fill = disease_rate)) +
geom_tile() +
scale_fill_gradient(low = "white", high = "red") +
theme_minimal() +
labs(title = "Disease Incidence by Sex and Alcohol Consumption",
x = "Sex",
y = "Alcohol Consumption Level",
fill = "Disease Rate") +
geom_text(aes(label = sprintf("n=%d", n)), color = "black", size = 3)
ggplotly(figure_2)A heatmap analysis revealed an inverse relationship between alcohol consumption and disease incidence. Non-drinkers exhibited the highest disease rates, with males outnumbering females. In contrast, participants with very high alcohol consumption had the lowest disease rates. Across all categories, females demonstrated slightly higher disease rates than males.
Blood Pressure and Age Trends
# Data Cleaning
disease_heat1 <- ukb_data %>%
filter(!is.na(age), !is.na(bp_cat), !is.na(nafld_outcome)) %>%
mutate(
bp_cat = as_factor(bp_cat),
age_group = cut(age,
breaks = c(30, 40, 50, 60, 70, 80),
labels = c("30-40", "40-50", "50-60", "60-70", "70-80"),
include.lowest = TRUE)
) %>%
group_by(age_group, bp_cat) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
# Create heatmap
figure_4 <- ggplot(disease_heat1, aes(x = age_group, y = bp_cat, fill = disease_rate)) +
geom_tile() +
scale_fill_gradient(low = "white", high = "red") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45)) +
labs(title = "Disease Incidence by Age and Blood Pressure Category",
x = "Age Groups",
y = "BP Category",
fill = "Disease Rate") +
geom_text(aes(label = sprintf("n=%d", n)), color = "black", size = 3)
ggplotly(figure_4)We observed that disease incidence increased significantly with higher blood pressure, particularly in individuals aged 60 and above. Younger age groups (30-40) showed low disease incidence even at higher blood pressure levels, while middle-aged and older groups demonstrated steeper increases. The largest sample sizes were concentrated in moderate blood pressure categories (BP=40-60).
Income and Diet Quality
color_palette <- colorRampPalette(c("lightblue", "darkblue"))(length(unique(ukb_data$diet_quality)))
figure_4 <- ukb_data %>%
filter(!is.na(income) & !is.na(diet_quality)) %>%
ggplot(aes(x = as.factor(income), fill = as.factor(diet_quality))) +
geom_bar(position = "stack") +
theme_minimal() +
scale_fill_manual(values = color_palette) +
labs(x = "Income Levels",
y = "Count",
fill = "Diet Quality",
title = "Income vs Diet Quality") +
theme(legend.position = "bottom")
ggplotly(figure_4)Diet quality showed a positive correlation with income levels. Participants in the highest income bracket had the highest proportions of top diet quality, while those in the lowest income bracket exhibited the poorest diet quality. Middle-income groups displayed more balanced distributions.
Sleep Duration Across Age Groups
figure_5 <- ukb_data %>%
filter(!is.na(age) & !is.na(sleep_hour)) %>%
mutate(age_group = cut(age, breaks = seq(10, 100, by = 10), right = FALSE,
labels = paste(seq(10, 90, by = 10), "-",
seq(19, 99, by = 10), sep = ""))) %>%
ggplot(aes(x = age_group, y = sleep_hour, fill = age_group)) +
geom_boxplot() +
theme_minimal() +
labs(x = "Age Groups",
y = "Sleep Hours per Night",
title = "Sleep Hours per Night by Age Groups",
fill = "Age Groups") +
scale_fill_manual(values = c("lightgreen", "lightpink", "lightblue",
"lightcoral", "lightgoldenrod"))
figure_5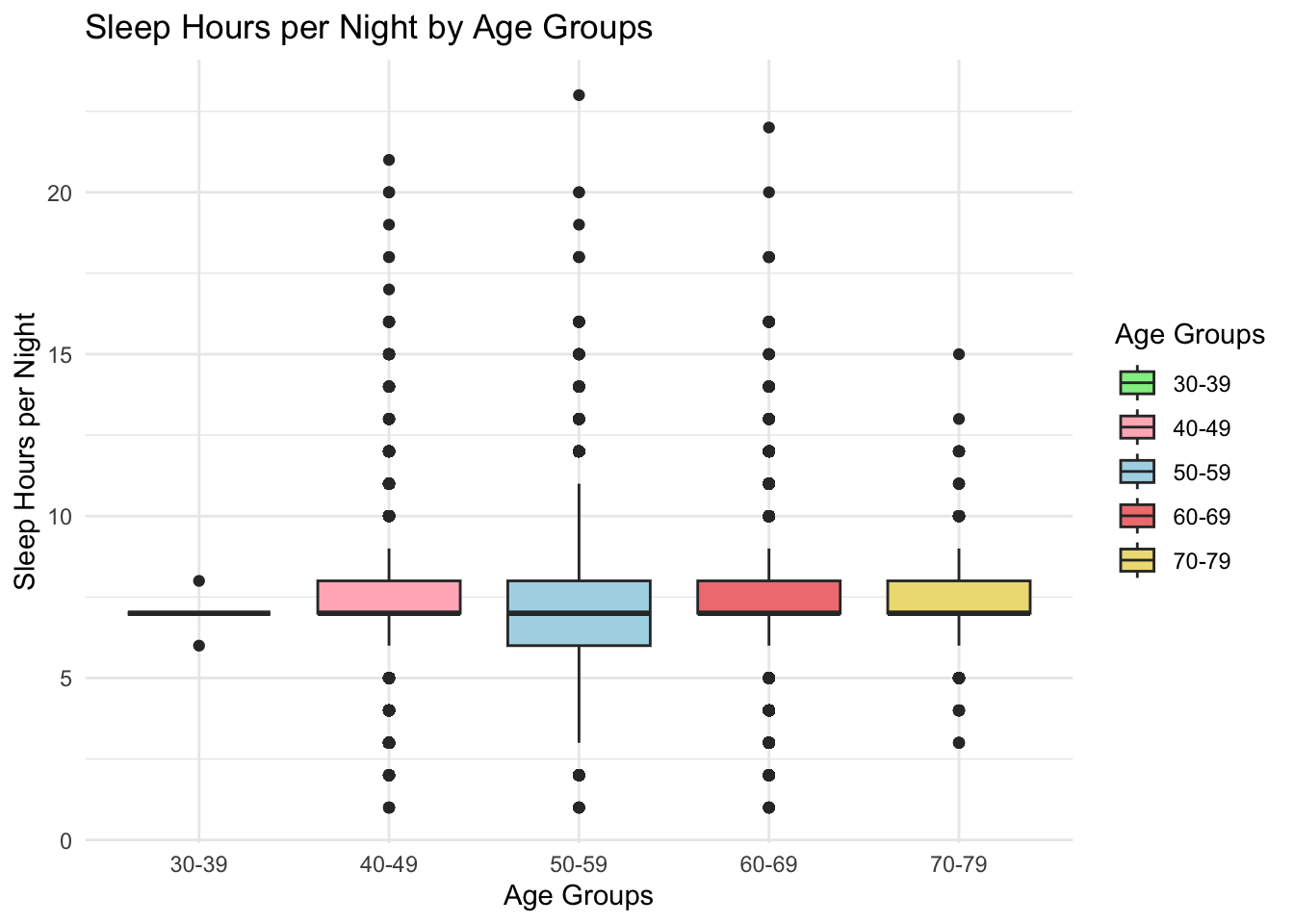
Sleep duration varied slightly by age. Younger participants (30-39) had the shortest median sleep duration and the highest prevalence of short sleep (<6 hours). Older age groups (60-69, 70-79) displayed greater variability, with outliers on both ends of the sleep duration spectrum, suggesting more diverse sleep patterns in older adults.
Exposure variables and disease outcomes
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_lmr)) %>%
summarize(
q25 = quantile(expo_lmr, 0.25),
median = median(expo_lmr),
q75 = quantile(expo_lmr, 0.75))
figure_6 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_lmr)) %>%
mutate(
lower_bound = quantile(expo_lmr, 0.25) - 1.5 * IQR(expo_lmr),
upper_bound = quantile(expo_lmr, 0.75) + 1.5 * IQR(expo_lmr)
) %>%
filter(expo_lmr >= lower_bound & expo_lmr <= upper_bound) %>%
ggplot(aes(x = expo_lmr, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte-to-Monocyte Ratio (LMR)",
y = "Density",
title = "Density Plot of expo_lmr by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_sii)) %>%
summarize(
q25 = quantile(expo_sii, 0.25),
median = median(expo_sii),
q75 = quantile(expo_sii, 0.75)
)
figure_7 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_sii)) %>%
mutate(
lower_bound = quantile(expo_sii, 0.25) - 1.5 * IQR(expo_sii),
upper_bound = quantile(expo_sii, 0.75) + 1.5 * IQR(expo_sii)
) %>%
filter(expo_sii >= lower_bound & expo_sii <= upper_bound) %>%
ggplot(aes(x = expo_sii, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Systemic Immune-Inflammation Index (SII)",
y = "Density",
title = "Density Plot of expo_sii by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_npar)) %>%
summarize(
q25 = quantile(expo_npar, 0.25),
median = median(expo_npar),
q75 = quantile(expo_npar, 0.75)
)
figure_8 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_npar)) %>%
mutate(
lower_bound = quantile(expo_npar, 0.25) - 1.5 * IQR(expo_npar),
upper_bound = quantile(expo_npar, 0.75) + 1.5 * IQR(expo_npar)
) %>%
filter(expo_npar >= lower_bound & expo_npar <= upper_bound) %>%
ggplot(aes(x = expo_npar, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophil-to-Albumin Ratio (NPAR)",
y = "Density",
title = "Density Plot of expo_npar by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
figure_9 <-ukb_data %>%
filter(!is.na(expo_nps)) %>%
group_by(nafld_outcome, expo_nps) %>%
summarize(count = n()) %>%
ungroup() %>%
mutate(total_count = sum(count),
percentage = count / total_count * 100) %>%
ggplot(aes(x = as.factor(expo_nps), y = percentage, fill = as.factor(nafld_outcome))) +
geom_bar(stat = "identity", position = position_dodge(width = 0.9), alpha = 0.8) +
geom_text(aes(label = paste0(round(percentage, 1), "%")),
position = position_dodge(width = 0.9), vjust = 0.75, size = 3) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Naples Prognostic Score (NPS)",
y = "Percentage of Total Population",
title = "Percentage of Total Population by NPS and NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
)
grid.arrange(figure_6, figure_7, figure_8, figure_9, nrow = 2, ncol = 2)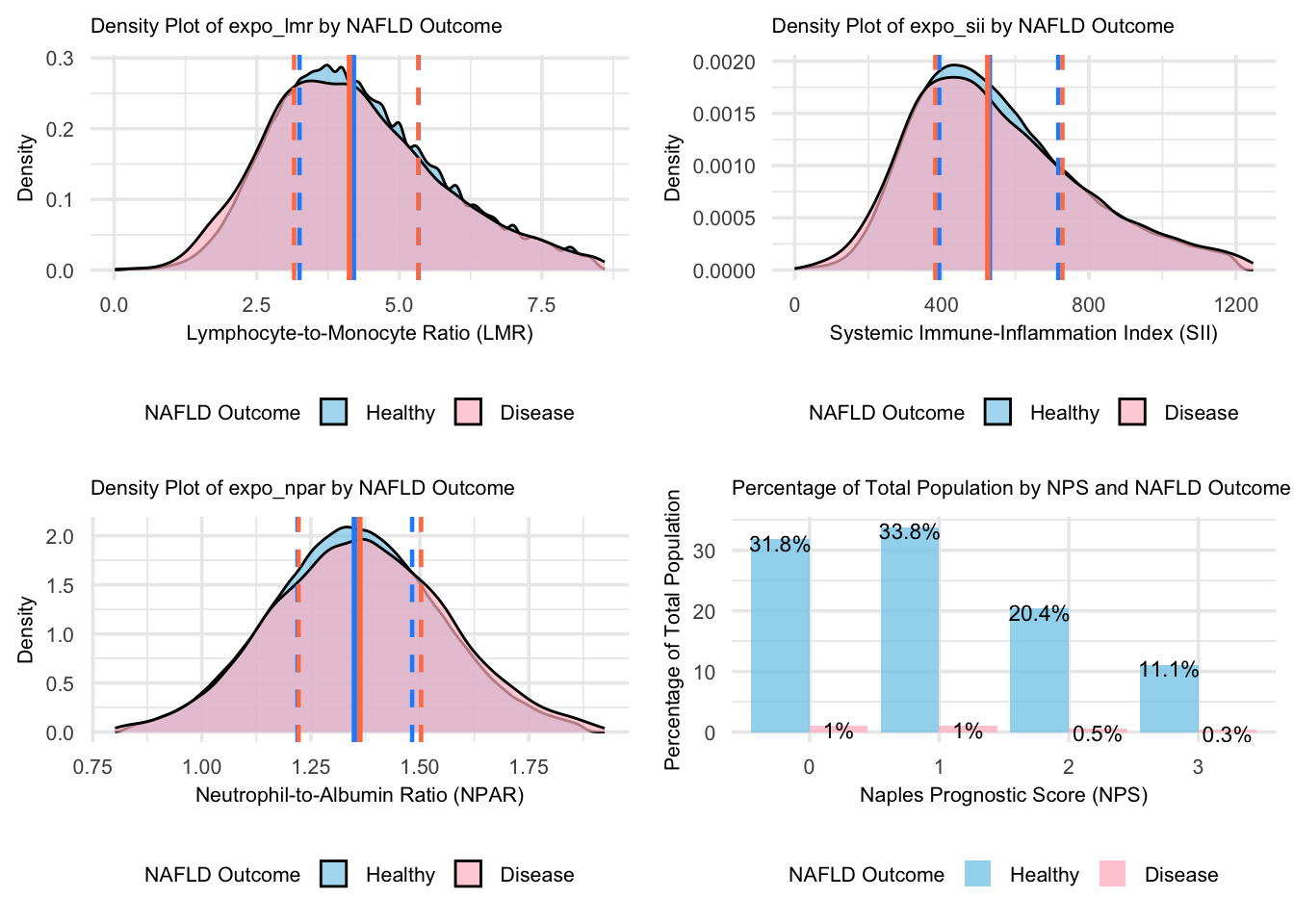
For the continuous biomarkers (LMR, SII, and NPAR), the density plots show consistently lower concentrations of values in the NAFLD group compared to the Healthy group, indicating a complex immune-inflammatory profile in NAFLD pathogenesis. The lower density peaks in LMR reflect more dispersed immune balance patterns, while decreased density in SII suggests more variable inflammatory responses. Similarly, the broader distribution of NPAR in NAFLD may indicate more heterogeneous liver function and neutrophil activity patterns. The vertical lines in each density plot represent the quartiles (Q1=25th percentile, Q2=median, and Q3=75th percentile), providing a clear visualization of the data distribution. The close median values and significant distribution overlap suggest limited discriminative power for these biomarkers alone in distinguishing NAFLD from healthy individuals. The NPS distribution shows higher proportions of healthy individuals across all scores, particularly in lower-risk categories (0-1), highlighting that while NAFLD alters immune-inflammatory profiles, this does not necessarily translate into higher prognostic risks.
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_lmr)) %>%
summarize(
q25 = quantile(expo_lmr, 0.25),
median = median(expo_lmr),
q75 = quantile(expo_lmr, 0.75))
figure_10 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_lmr)) %>%
mutate(
lower_bound = quantile(expo_lmr, 0.25) - 1.5 * IQR(expo_lmr),
upper_bound = quantile(expo_lmr, 0.75) + 1.5 * IQR(expo_lmr)
) %>%
filter(expo_lmr >= lower_bound & expo_lmr <= upper_bound) %>%
ggplot(aes(x = expo_lmr, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte-to-Monocyte Ratio (LMR)",
y = "Density",
title = "Density Plot of expo_lmr by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_sii)) %>%
summarize(
q25 = quantile(expo_sii, 0.25),
median = median(expo_sii),
q75 = quantile(expo_sii, 0.75)
)
figure_11 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_sii)) %>%
mutate(
lower_bound = quantile(expo_sii, 0.25) - 1.5 * IQR(expo_sii),
upper_bound = quantile(expo_sii, 0.75) + 1.5 * IQR(expo_sii)
) %>%
filter(expo_sii >= lower_bound & expo_sii <= upper_bound) %>%
ggplot(aes(x = expo_sii, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Systemic Immune-Inflammation Index (SII)",
y = "Density",
title = "Density Plot of expo_sii by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_npar)) %>%
summarize(
q25 = quantile(expo_npar, 0.25),
median = median(expo_npar),
q75 = quantile(expo_npar, 0.75)
)
figure_12 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_npar)) %>%
mutate(
lower_bound = quantile(expo_npar, 0.25) - 1.5 * IQR(expo_npar),
upper_bound = quantile(expo_npar, 0.75) + 1.5 * IQR(expo_npar)
) %>%
filter(expo_npar >= lower_bound & expo_npar <= upper_bound) %>%
ggplot(aes(x = expo_npar, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophil-to-Albumin Ratio (NPAR)",
y = "Density",
title = "Density Plot of expo_npar by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
figure_13 <- ukb_data %>%
filter(!is.na(expo_nps)) %>%
group_by(cirrho_outcome, expo_nps) %>%
summarize(count = n()) %>%
ungroup() %>%
mutate(total_count = sum(count),
percentage = count / total_count * 100) %>%
ggplot(aes(x = as.factor(expo_nps), y = percentage, fill = as.factor(cirrho_outcome))) +
geom_bar(stat = "identity", position = position_dodge(width = 0.9), alpha = 0.8) +
geom_text(aes(label = paste0(round(percentage, 1), "%")),
position = position_dodge(width = 0.9), vjust = 0.5, size = 3) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Naples Prognostic Score (NPS)",
y = "Percentage of Total Population",
title = "Percentage of Total Population by NPS and Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
)
grid.arrange(figure_10, figure_11, figure_12, figure_13, nrow = 2, ncol = 2)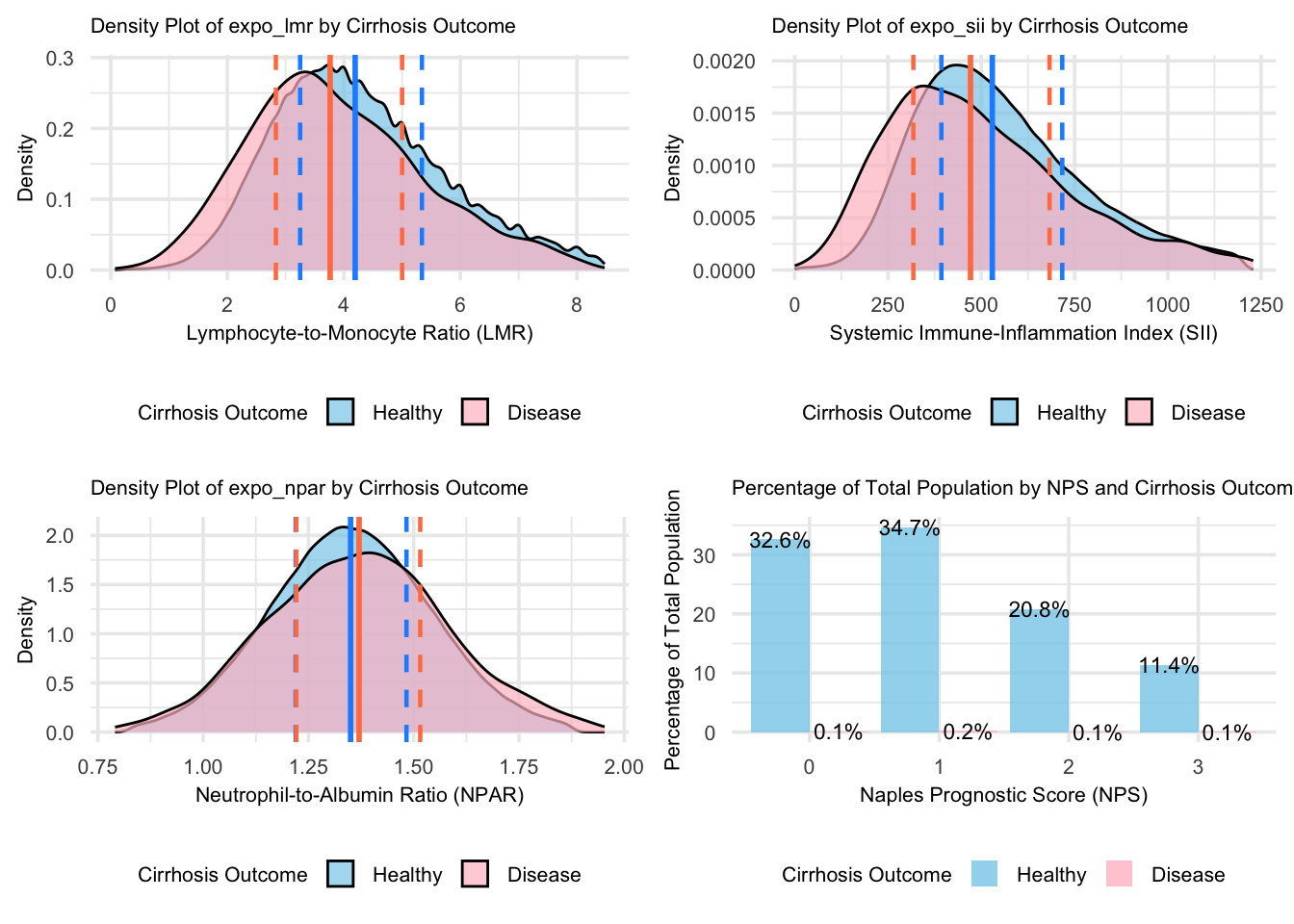
For the Cirrhosis group, the density plots reveal distinct distribution patterns compared to the Healthy group: LMR and SII show lower peak heights in cirrhosis, indicating more dispersed values and suggesting variable immune and inflammatory states, while NPAR shows shifted peak concentrations, reflecting altered liver function patterns. The height and shape of the density curves suggest more concentrated value distributions in healthy individuals for LMR and SII, indicating more consistent immune-inflammatory states in health. Though the overlapping areas between curves indicate some shared value ranges, the different peak heights and positions reflect distinct disease characteristics. The NPS distribution further emphasizes this contrast, with higher density concentrations in healthy individuals across risk categories.
Blood parameters and disease outcomes
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_lympho)) %>%
summarize(
q25 = quantile(b_lympho, 0.25),
median = median(b_lympho),
q75 = quantile(b_lympho, 0.75))
figure_14 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_lympho)) %>%
mutate(
lower_bound = quantile(b_lympho, 0.25) - 1.5 * IQR(b_lympho),
upper_bound = quantile(b_lympho, 0.75) + 1.5 * IQR(b_lympho)
) %>%
filter(b_lympho >= lower_bound & b_lympho <= upper_bound) %>%
ggplot(aes(x = b_lympho, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte",
y = "Density",
title = "Density Plot of b_lympho by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_mono)) %>%
summarize(
q25 = quantile(b_mono, 0.25),
median = median(b_mono),
q75 = quantile(b_mono, 0.75)
)
figure_15 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_mono)) %>%
mutate(
lower_bound = quantile(b_mono, 0.25) - 1.5 * IQR(b_mono),
upper_bound = quantile(b_mono, 0.75) + 1.5 * IQR(b_mono)
) %>%
filter(b_mono >= lower_bound & b_mono <= upper_bound) %>%
ggplot(aes(x = b_mono, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Monocytes",
y = "Density",
title = "Density Plot of b_mono by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_neutro)) %>%
summarize(
q25 = quantile(b_neutro, 0.25),
median = median(b_neutro),
q75 = quantile(b_neutro, 0.75)
)
figure_16 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_neutro)) %>%
mutate(
lower_bound = quantile(b_neutro, 0.25) - 1.5 * IQR(b_neutro),
upper_bound = quantile(b_neutro, 0.75) + 1.5 * IQR(b_neutro)
) %>%
filter(b_neutro >= lower_bound & b_neutro <= upper_bound) %>%
ggplot(aes(x = b_neutro, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophils",
y = "Density",
title = "Density Plot of b_neutro by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_plate)) %>%
summarize(
q25 = quantile(b_plate, 0.25),
median = median(b_plate),
q75 = quantile(b_plate, 0.75)
)
figure_17 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_plate)) %>%
mutate(
lower_bound = quantile(b_plate, 0.25) - 1.5 * IQR(b_plate),
upper_bound = quantile(b_plate, 0.75) + 1.5 * IQR(b_plate)
) %>%
filter(b_plate >= lower_bound & b_plate <= upper_bound) %>%
ggplot(aes(x = b_plate, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Platelets",
y = "Density",
title = "Density Plot of b_plate by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
grid.arrange(figure_14, figure_15, figure_16, figure_17, nrow = 2, ncol = 2)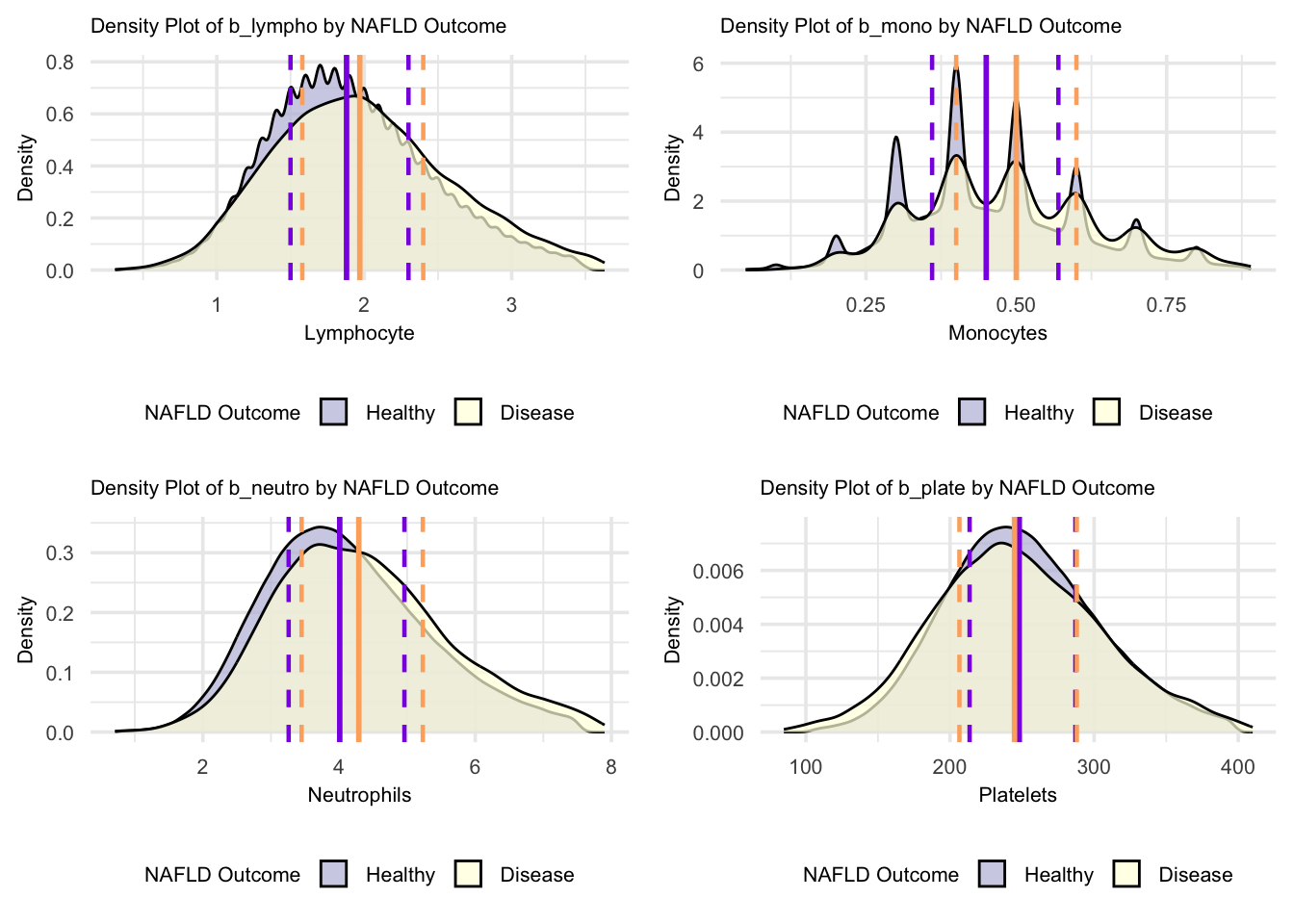
For these blood parameters (lymphocytes, monocytes, neutrophils, and platelets), the density plots reveal distinct distribution patterns. The monocyte plot shows multiple sharp peaks, indicating distinct clustered values in both groups. Lymphocyte and neutrophil distributions show bell-shaped curves with considerable overlap but different peak heights, suggesting concentrated value ranges in both healthy and NAFLD states. Platelet distributions show broader, more spread-out patterns with similar shapes between groups, indicating more variable platelet counts in both populations. These density patterns suggest that while the value ranges overlap, the concentration of measurements differs between healthy and NAFLD states, with each parameter showing unique distribution characteristics.
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_lympho)) %>%
summarize(
q25 = quantile(b_lympho, 0.25),
median = median(b_lympho),
q75 = quantile(b_lympho, 0.75))
figure_18 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_lympho)) %>%
mutate(
lower_bound = quantile(b_lympho, 0.25) - 1.5 * IQR(b_lympho),
upper_bound = quantile(b_lympho, 0.75) + 1.5 * IQR(b_lympho)
) %>%
filter(b_lympho >= lower_bound & b_lympho <= upper_bound) %>%
ggplot(aes(x = b_lympho, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte",
y = "Density",
title = "Density Plot of b_lympho by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_mono)) %>%
summarize(
q25 = quantile(b_mono, 0.25),
median = median(b_mono),
q75 = quantile(b_mono, 0.75)
)
figure_19 <-ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_mono)) %>%
mutate(
lower_bound = quantile(b_mono, 0.25) - 1.5 * IQR(b_mono),
upper_bound = quantile(b_mono, 0.75) + 1.5 * IQR(b_mono)
) %>%
filter(b_mono >= lower_bound & b_mono <= upper_bound) %>%
ggplot(aes(x = b_mono, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Monocytes",
y = "Density",
title = "Density Plot of b_mono by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_neutro)) %>%
summarize(
q25 = quantile(b_neutro, 0.25),
median = median(b_neutro),
q75 = quantile(b_neutro, 0.75)
)
figure_20 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_neutro)) %>%
mutate(
lower_bound = quantile(b_neutro, 0.25) - 1.5 * IQR(b_neutro),
upper_bound = quantile(b_neutro, 0.75) + 1.5 * IQR(b_neutro)
) %>%
filter(b_neutro >= lower_bound & b_neutro <= upper_bound) %>%
ggplot(aes(x = b_neutro, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophils",
y = "Density",
title = "Density Plot of b_neutro by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_plate)) %>%
summarize(
q25 = quantile(b_plate, 0.25),
median = median(b_plate),
q75 = quantile(b_plate, 0.75)
)
figure_21 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_plate)) %>%
mutate(
lower_bound = quantile(b_plate, 0.25) - 1.5 * IQR(b_plate),
upper_bound = quantile(b_plate, 0.75) + 1.5 * IQR(b_plate)
) %>%
filter(b_plate >= lower_bound & b_plate <= upper_bound) %>%
ggplot(aes(x = b_plate, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Platelets",
y = "Density",
title = "Density Plot of b_plate by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
grid.arrange(figure_18, figure_19, figure_20, figure_21, nrow = 2, ncol = 2)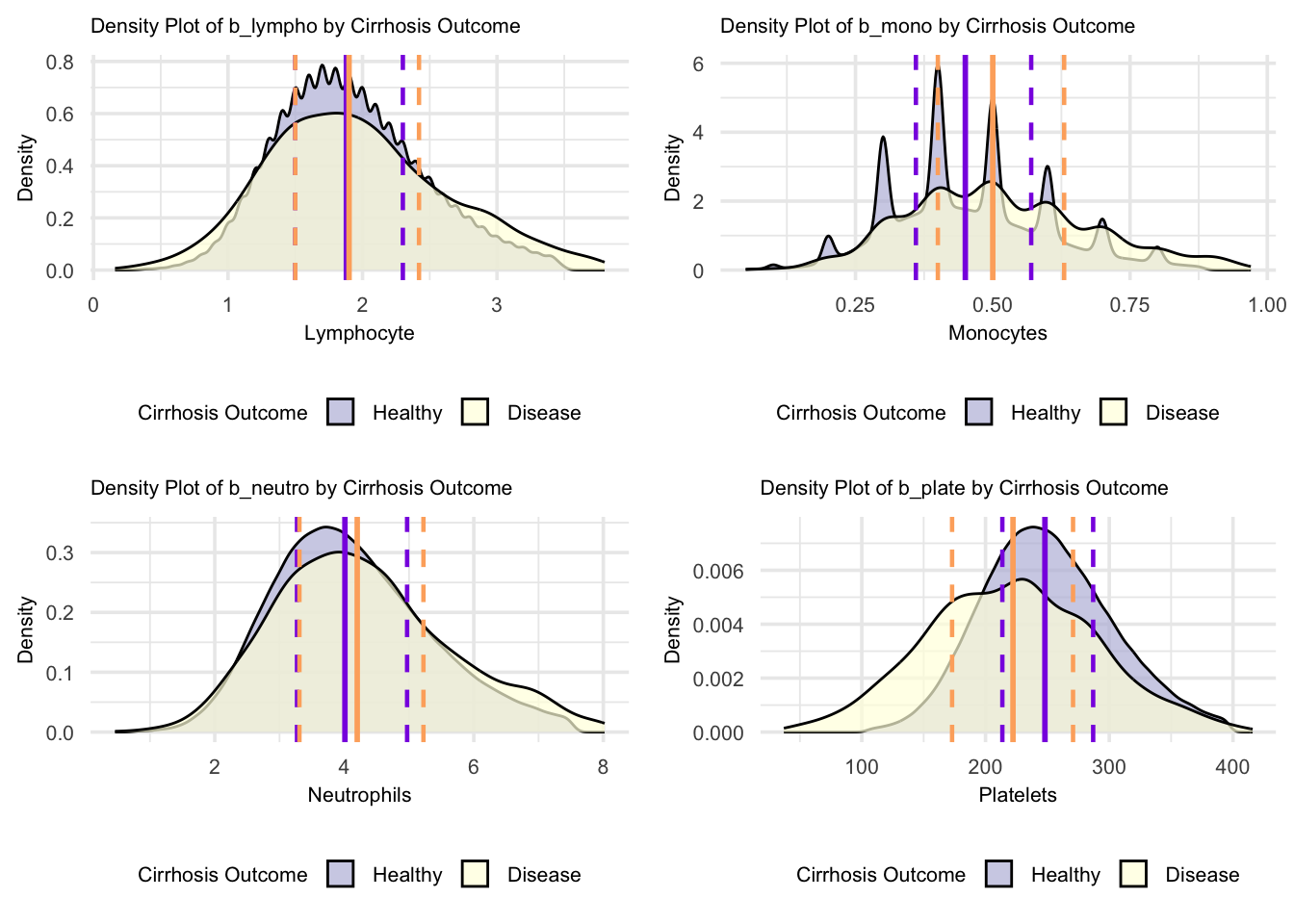
For these blood parameters, the density plots demonstrate distinctive patterns between Healthy and Cirrhosis groups. The lymphocyte distribution shows similar peak heights, while monocytes display multiple sharp peaks indicating distinct value clusters in both groups. Neutrophil distributions show broader patterns with different peak positions between groups, suggesting shifted but overlapping value ranges. The platelet distribution shows the most striking difference, with a notably higher and more concentrated peak in the healthy group compared to a lower, more dispersed distribution in cirrhosis, indicating more consistent platelet counts in health versus more variable counts in disease. Looking at the specific median lines, lymphocytes show almost identical values between healthy and cirrhosis groups, while monocytes and neutrophils demonstrate higher median values in the cirrhosis group compared to healthy individuals. In contrast, platelets show a notably higher median in the healthy group compared to cirrhosis, with the most pronounced difference among all parameters, likely reflecting the impact of portal hypertension in cirrhosis. Distribution overlaps exist between groups across all parameters, though the platelet parameter shows the clearest separation between healthy and cirrhosis populations.
Survival Analysis
View Full Survival Analysis Steps on GitHub
Methodology
Cox proportional hazards models will be employed to investigate the association between exposure categories and survival outcomes. In this study, we will conduct survival analysis to evaluate the relationships between baseline inflammatory and metabolic exposure categories and the risks of developing non-alcoholic fatty liver disease (NAFLD) and cirrhosis. Survival analysis is a statistical approach designed to analyze time-to-event data, where the survival time \(T\) (time until the occurrence of the event of interest) and the censoring time \(C\) (time at which an observation is no longer tracked) are observed. The observed survival time, denoted as:
\[Y = min(T, C)\]
The censoring indicator \(\delta\) is defined as: \[ \delta = \begin{cases} 1 & \text{if } T \leq C \\ 0 & \text{if } T > C \end{cases} \] Here, \(\delta = 1\) indicates the event was observed, and \(\delta = 0\) signifies that censoring occurred.
By modeling survival times and event occurrences, we aim to quantify the impact of different exposure categories on the hazard of developing NAFLD and cirrhosis, adjusting for relevant covariates.
General Form of the Cox Proportional Hazards Model
We applied the Cox proportional hazards model to evaluate how various covariates influence the hazard rate \(h(t)\), which represents the instantaneous risk of the event occurring at time \(t\). The hazard function is defined as:
\[h(t) = h_0(t) \exp(x_1\beta_1 + x_2\beta_2 + \dots + x_k\beta_k)\]
Where:
\(h(t)\): Hazard function for time t,
\(h_0(t)\): Baseline hazard function,
\(x_i\): Covariates (e.g., age, sex, inflammatory markers, socioeconomic factors),
\(\beta_i\): Regression coefficients for covariates.
Baseline Characteristics Table
The baseline characteristics are stratified by each exposure category to identify differences in fundamental features across exposure groups and to provide a reference for constructing the Cox proportional hazards model (see Appendix for more detailed tables and explanations).
Baseline_expo_lmr_cat_results <- read_csv("./csv/Baseline_expo_lmr_cat.csv")
Baseline_expo_lmr_cat_cleaned <- Baseline_expo_lmr_cat_results %>%
select(-test) %>%
rename(
` ` = ...1,
Level1 = `1`,
Level2 = `2`,
Level3 = `3`
)
knitr::kable(
Baseline_expo_lmr_cat_cleaned,
digits = 2,
caption = "Baseline Characteristics by LMR Exposure Categories",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Overall | Level1 | Level2 | Level3 | p | |
|---|---|---|---|---|---|
| n | 413146 | 105227 | 204634 | 103279 | NA |
| sex = 2 (%) | 190849 (46.2) | 69149 (65.7) | 93068 (45.5) | 28629 (27.7) | <0.001 |
| age (mean (SD)) | 56.54 (8.09) | 57.70 (8.07) | 56.32 (8.10) | 55.79 (7.96) | <0.001 |
| income (%) | NA | NA | NA | NA | <0.001 |
| 1 | 40844 (9.9) | 9912 (9.4) | 20134 (9.9) | 10798 (10.5) | NA |
| 2 | 17370 (4.2) | 3874 (3.7) | 8290 (4.1) | 5206 (5.1) | NA |
| 3 | 79972 (19.4) | 21680 (20.7) | 37894 (18.6) | 20398 (19.8) | NA |
| 4 | 89874 (21.8) | 23863 (22.8) | 44107 (21.6) | 21902 (21.3) | NA |
| 5 | 92455 (22.4) | 23555 (22.5) | 46634 (22.8) | 22263 (21.6) | NA |
| 6 | 72283 (17.5) | 17494 (16.7) | 37205 (18.2) | 17583 (17.1) | NA |
| 7 | 19249 (4.7) | 4513 (4.3) | 9872 (4.8) | 4864 (4.7) | NA |
| townsend (mean (SD)) | -1.33 (3.08) | -1.39 (3.04) | -1.39 (3.04) | -1.14 (3.18) | <0.001 |
| total_met (mean (SD)) | 1619.08 (2015.70) | 1645.29 (2084.82) | 1625.33 (2008.34) | 1578.85 (1955.34) | <0.001 |
| diet_quality (mean (SD)) | 3.29 (1.57) | 3.12 (1.57) | 3.31 (1.57) | 3.42 (1.56) | <0.001 |
| sleep_hour (mean (SD)) | 7.15 (1.10) | 7.17 (1.12) | 7.15 (1.09) | 7.14 (1.13) | <0.001 |
| smoke_cat (%) | NA | NA | NA | NA | <0.001 |
| 1 | 32255 (7.8) | 6743 (6.4) | 15030 (7.3) | 10481 (10.1) | NA |
| 2 | 11334 (2.7) | 2843 (2.7) | 5572 (2.7) | 2919 (2.8) | NA |
| 3 | 96062 (23.3) | 27859 (26.5) | 47078 (23.0) | 21122 (20.5) | NA |
| 4 | 47431 (11.5) | 12141 (11.5) | 23827 (11.6) | 11463 (11.1) | NA |
| 5 | 60211 (14.6) | 15267 (14.5) | 30436 (14.9) | 14507 (14.0) | NA |
| 6 | 165853 (40.1) | 40374 (38.4) | 82691 (40.4) | 42787 (41.4) | NA |
| total_alcohol (mean (SD)) | 12.55 (15.17) | 15.16 (17.37) | 12.50 (14.77) | 9.98 (12.92) | <0.001 |
| non_hdl (mean (SD)) | 359.49 (78.74) | 345.15 (77.61) | 361.10 (77.88) | 371.09 (79.37) | <0.001 |
| tg (mean (SD)) | 92.17 (22.17) | 92.24 (22.59) | 91.90 (21.60) | 92.66 (22.85) | <0.001 |
| bp_cat (mean (SD)) | 49.88 (28.78) | 47.57 (28.26) | 50.05 (28.71) | 51.91 (29.25) | <0.001 |
This table provides the baseline characteristics of participants categorized by Lymphocyte-to-Monocyte Ratio (LMR) levels (Level 1, Level 2, Level 3). It includes demographic (e.g., age, sex), socioeconomic (e.g., income, Townsend index), lifestyle (e.g., smoking, alcohol intake), and clinical markers (e.g., triglycerides, diet quality), highlighting significant differences across categories with p-values for most variables. These differences emphasize potential confounding variables that need adjustment in subsequent statistical models, such as Cox proportional hazards analysis.
Cox Proportional Hazards Models
Model 1
\[ h(t) = h_0(t) \exp\left( \beta_{\text{expo}} \times x_{expo} + \beta_{\text{age}} \times x_{age} + \beta_{\text{sex}} \times x_{sex} \right) \]
Model 2
\[ \begin{aligned} h(t) = h_0(t) \exp\Big( & \beta_{\text{expo}} \times x_{expo} + \beta_{\text{age}} \times x_{age} + \beta_{\text{sex}} \times x_{sex} + \beta_{\text{income}} \times x_{income} \\ & + \beta_{\text{townsend}} \times x_{townsend} + \beta_{\text{total_met}} \times x_{\text{total_met}} + \beta_{\text{diet_quality}} \times x_{\text{diet_quality}} \\ & + \beta_{\text{sleep_hour}} \times x_{\text{sleep_hour}} + \beta_{\text{smoke_cat}} \times x_{\text{smoke_cat}} + \beta_{\text{total_alcohol}} \times x_{\text{total_alcohol}} \\ & + \beta_{\text{no_hdl}} \times x_{\text{no_hdl}} + \beta_{\text{tg}} \times x_{\text{tg}} + \beta_{\text{bp_cat}} \times x_{\text{bp_cat}} \Big) \end{aligned} \]
After fitting the Cox proportional hazards models, hazard ratios will be calculated for each exposure category, with the lowest exposure category serving as the reference. Statistical significance will be assessed using p-values, and dose-response relationships will be evaluated through p-for-trend tests. Results will be reported with 95% confidence intervals to convey precision. Survival durations will be computed based on either censored times or event times, with imputation applied where necessary to ensure complete datasets for analysis.
Cumulative Hazard Plots
Figure 1: Cumulative Hazard Plot for expo_lmr Categories in NAFLD Outcome
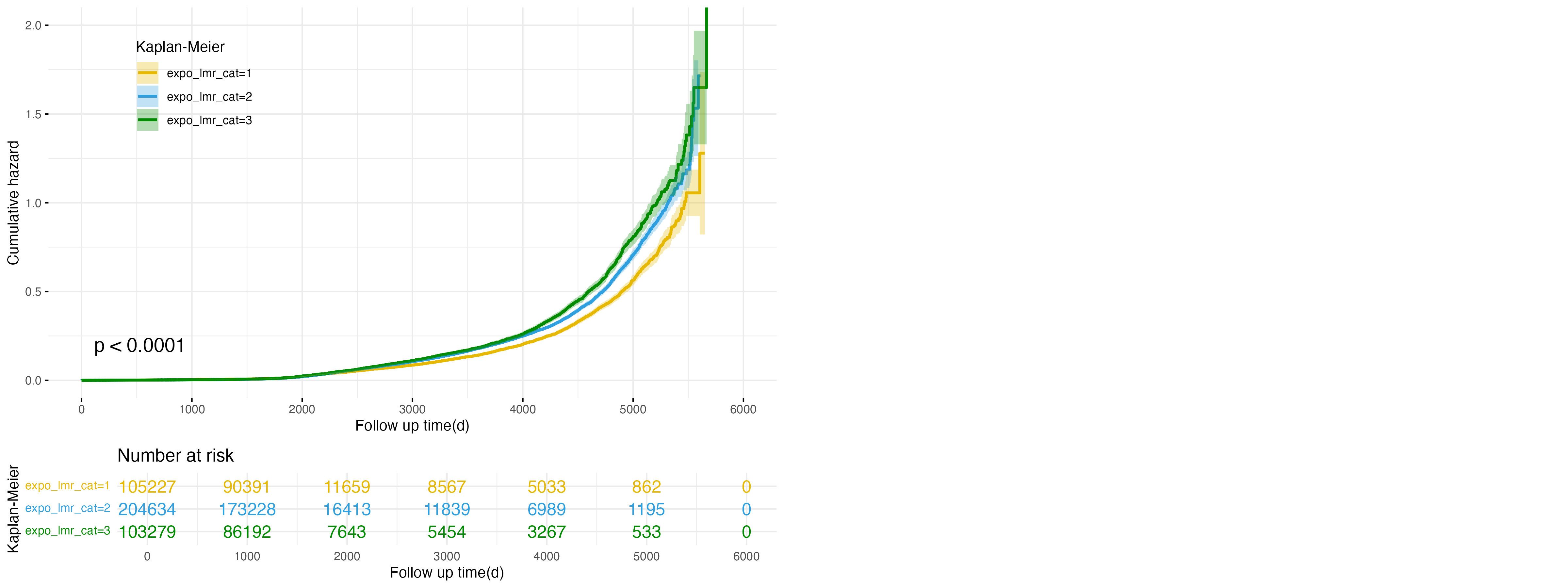
Figure 1 demonstrates the association between lymphocyte-to-monocyte ratio (LMR) exposure categories and the risk of developing non-alcoholic fatty liver disease (NAFLD). The high exposure group (expo_lmr_cat=3, green line) exhibits the highest cumulative hazard throughout the follow-up period, followed by the intermediate (expo_lmr_cat=2, blue line) and low exposure (expo_lmr_cat=1, yellow line) groups. These differences are statistically significant (p < 0.0001), indicating that higher LMR levels are associated with an increased risk of NAFLD. This suggests that elevated LMR, potentially indicative of an immune imbalance, may contribute to the progression of NAFLD.
Figure 2: Cumulative Hazard Plot for expo_lmr Categories in Cirrhosis Outcome
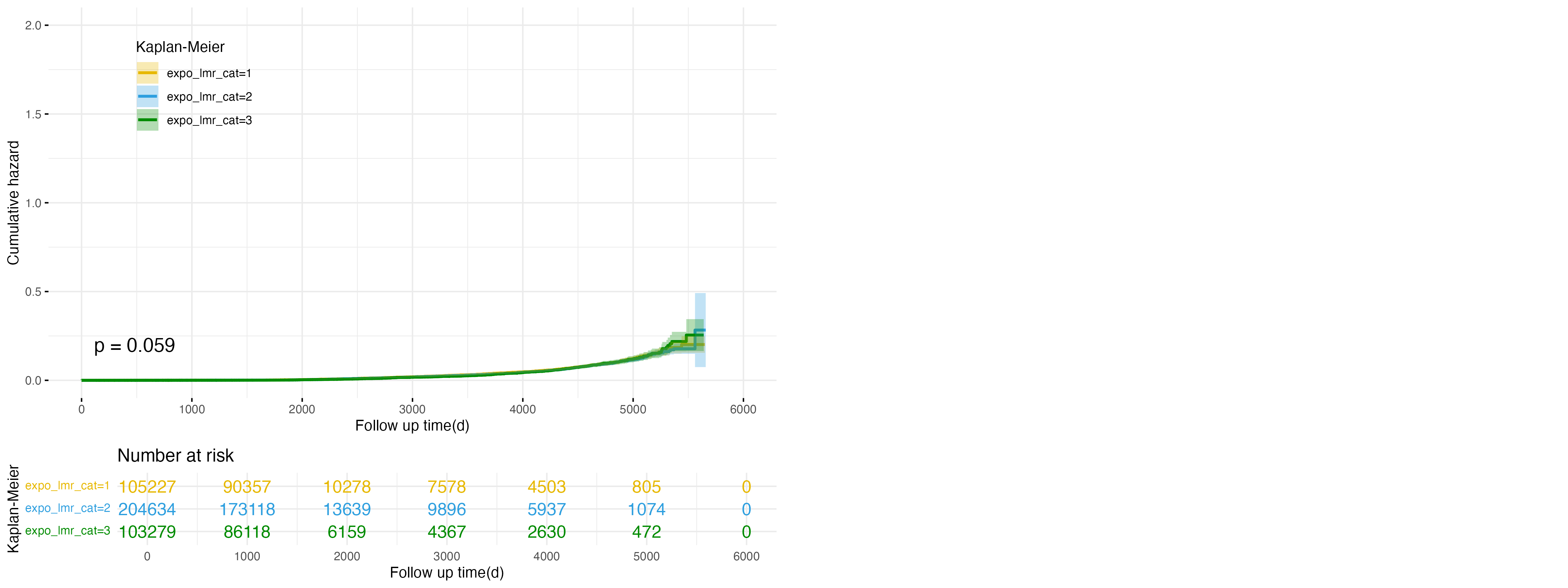
Expanding on the findings for NAFLD, Figure 2 evaluates the relationship between LMR exposure categories and the risk of developing cirrhosis. The high exposure group (expo_lmr_cat=3, green line) again demonstrates the highest cumulative hazard, followed by the intermediate (expo_lmr_cat=2, blue line) and low exposure (expo_lmr_cat=1, yellow line) groups. Although the separation of the curves becomes more evident after approximately 5,000 days, the differences are not statistically significant (p = 0.059). These findings suggest that higher LMR levels may also be associated with an increased risk of cirrhosis, though the evidence is weaker compared to NAFLD.
Figure 3: Cumulative Hazard Plot for SII Exposure in NAFLD Outcome
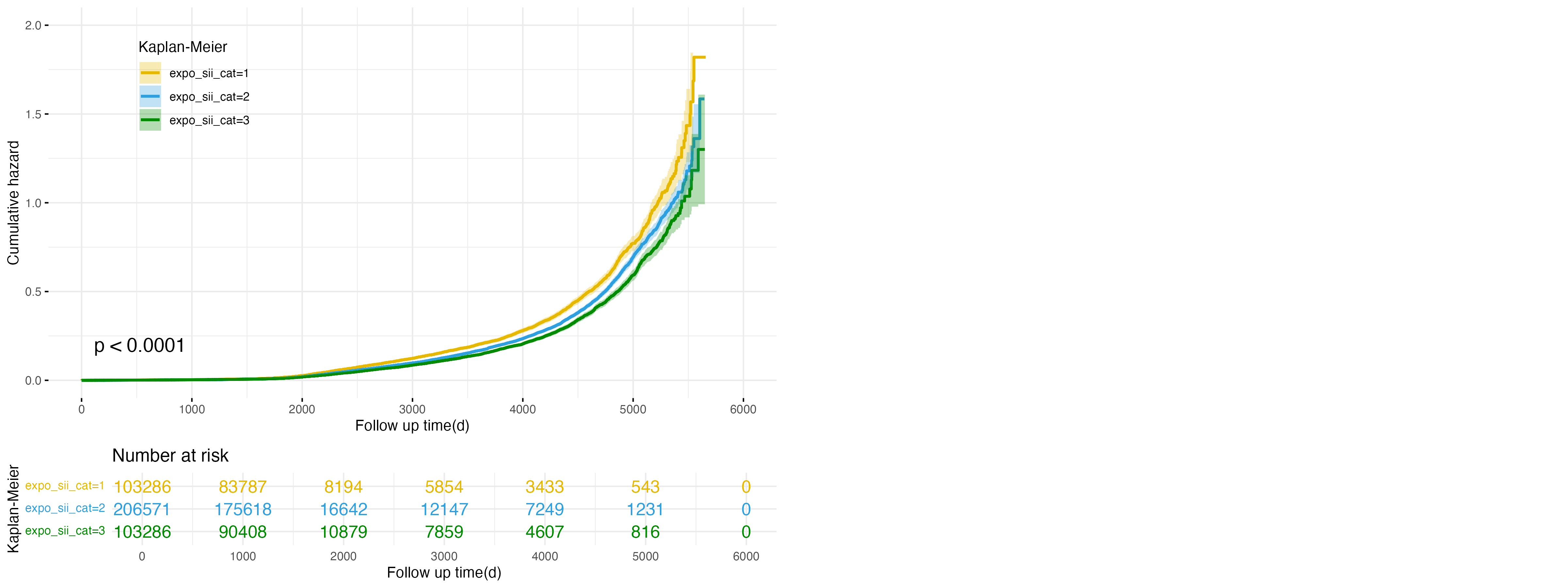
Building on the role of inflammatory markers, Figure 3 illustrates the relationship between systemic immune-inflammation index (SII) exposure categories and the risk of developing NAFLD. The low exposure group (expo_sii_cat=1, yellow line) exhibits the highest cumulative hazard, followed by the intermediate group (expo_sii_cat=2, blue line), while the high exposure group (expo_sii_cat=3, green line) shows the lowest cumulative hazard. These differences are statistically significant (p < 0.0001), indicating that higher SII levels are associated with a reduced risk of NAFLD. This finding highlights the complex role of systemic inflammation, where individuals with lower SII levels may have an elevated risk of NAFLD.
Figure 4: Cumulative Hazard Plot for SII Exposure in Cirrhosis Outcome
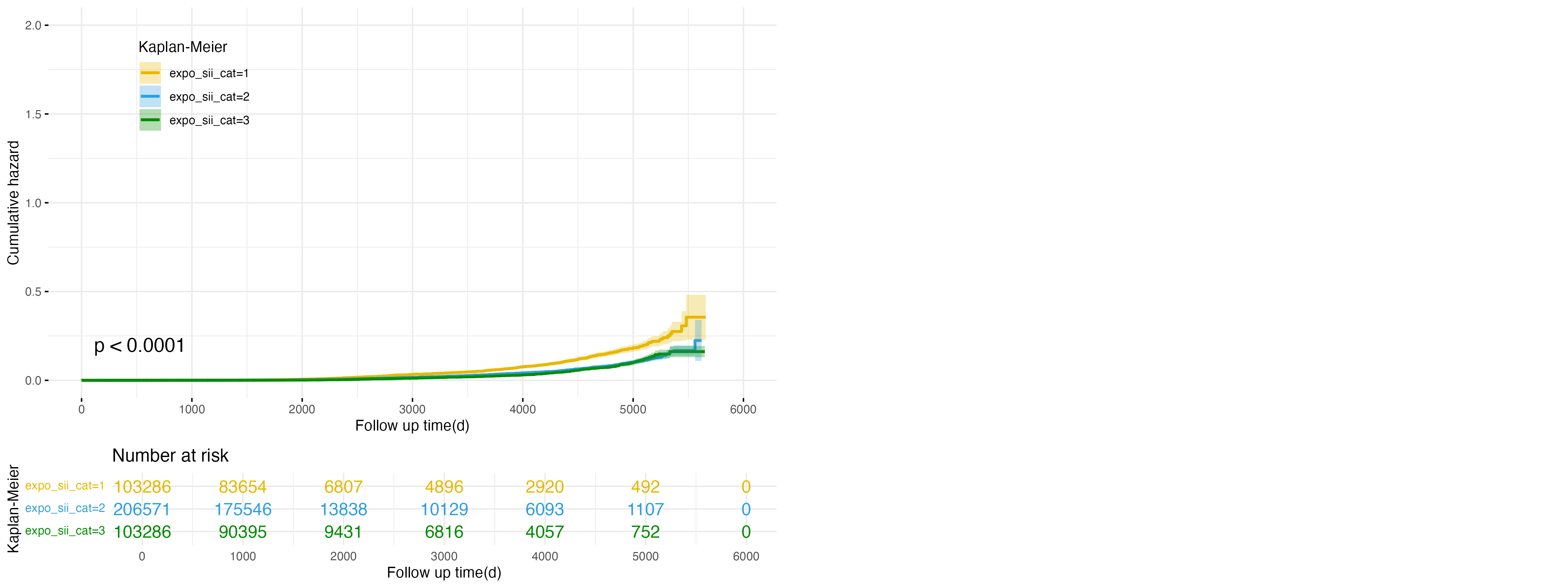
Expanding the analysis to cirrhosis, Figure 4 examines the association between SII exposure categories and cirrhosis risk. Similar to the trend observed in NAFLD, the low exposure group (expo_sii_cat=1, yellow line) shows the highest cumulative hazard, followed by the intermediate group (expo_sii_cat=2, blue line), while the high exposure group (expo_sii_cat=3, green line) consistently demonstrates the lowest cumulative hazard. These differences are statistically significant (p < 0.0001), suggesting that higher SII levels are associated with a reduced risk of cirrhosis. This pattern indicates that systemic inflammation, as reflected by lower SII levels, may play a role in cirrhosis development.
Figure 5: Cumulative Hazard Plot for NPAR Exposure in NAFLD Outcome
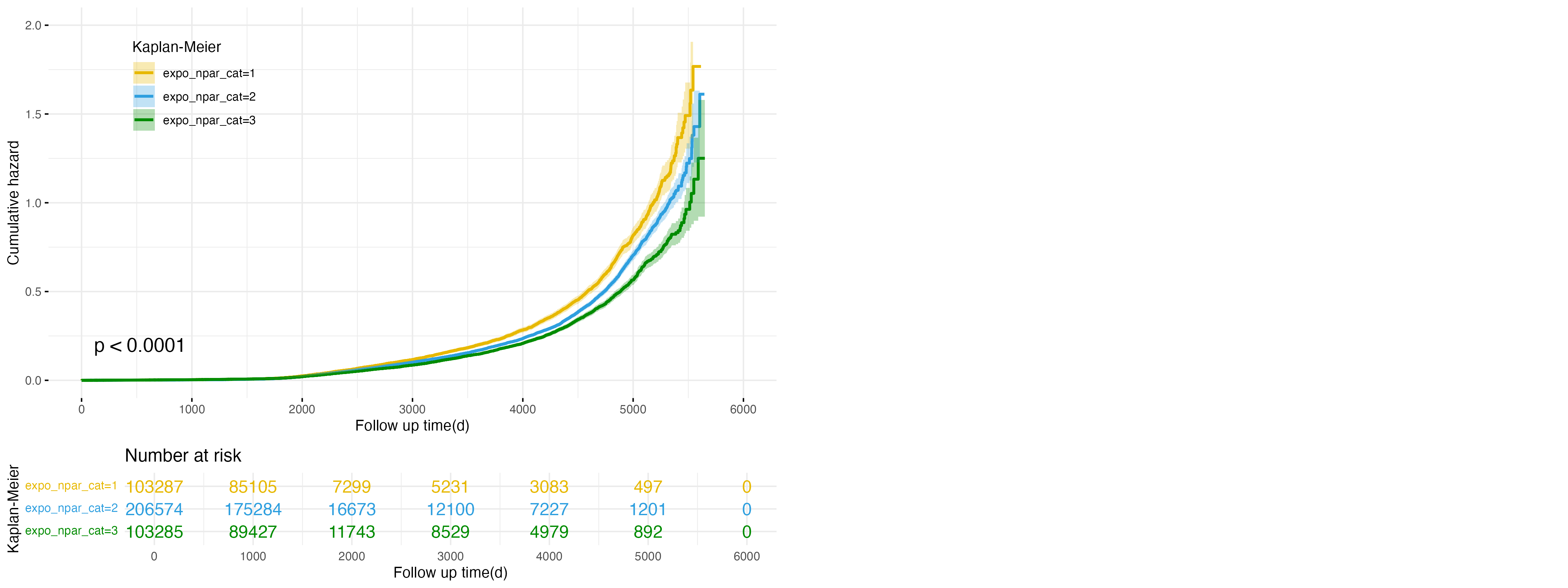
Further exploring inflammatory markers, Figure 5 illustrates the relationship between Neutrophil-Percentage-to-Albumin Ratio (NPAR) exposure categories and NAFLD risk. The low exposure group (expo_npar_cat=1, yellow line) exhibits the highest cumulative hazard, followed by the intermediate group (expo_npar_cat=2, blue line), while the high exposure group (expo_npar_cat=3, green line) demonstrates the lowest cumulative hazard. These differences are statistically significant (p < 0.0001), indicating that higher NPAR levels are associated with a reduced risk of NAFLD. These findings emphasize the potential protective role of higher NPAR levels against NAFLD progression.
Figure 6: Cumulative Hazard Plot for NPAR Exposure in Cirrhosis Outcome
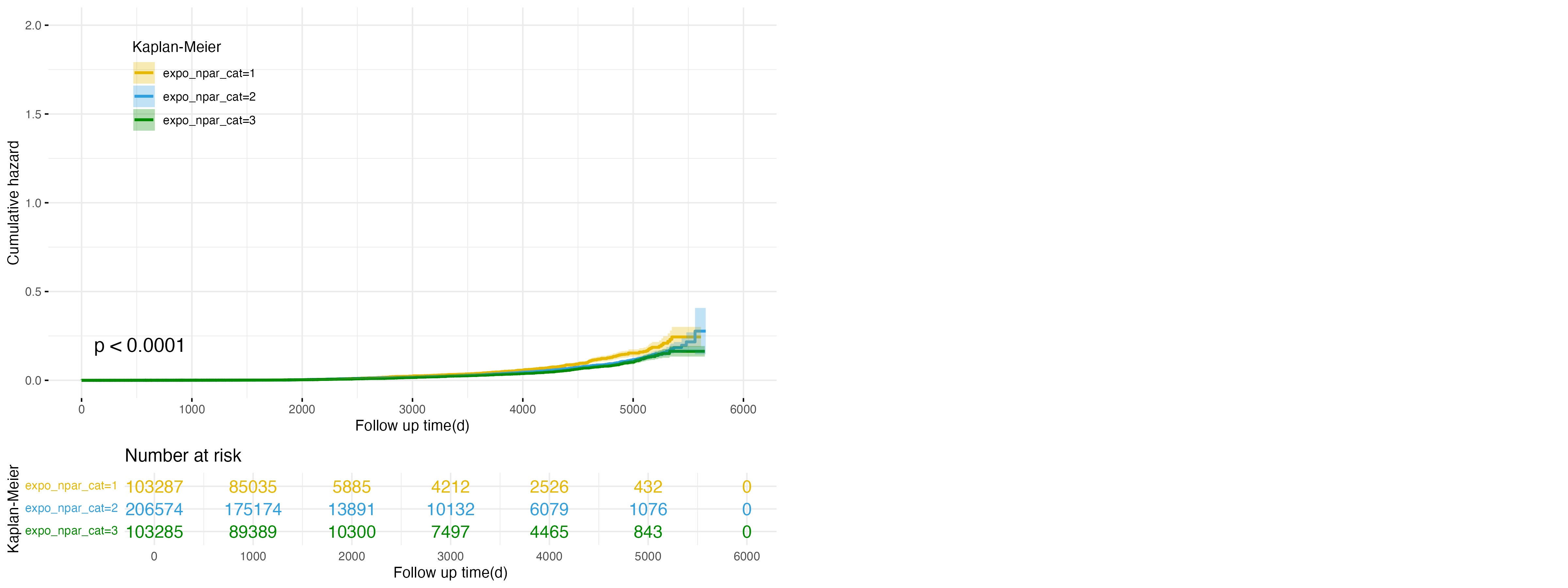
Finally, Figure 6 explores the relationship between NPAR exposure categories and the risk of cirrhosis. The low exposure group (expo_npar_cat=1, yellow line) shows the highest cumulative hazard, followed by the intermediate group (expo_npar_cat=2, blue line), while the high exposure group (expo_npar_cat=3, green line) consistently exhibits the lowest cumulative hazard. These differences are statistically significant (p < 0.0001), suggesting that higher NPAR levels are associated with a reduced risk of cirrhosis. This further supports the protective role of higher NPAR levels in mitigating cirrhosis risk, potentially through modulation of systemic inflammation.
Results for NAFLD Outcome
nafld_results <- read_csv("./csv/Main_cox_nafld.csv")
nafld_results_cleaned <- nafld_results %>%
select(-1, -lower_conf, -upper_conf) %>%
rename(
Model = Model,
`Exposure Category` = Category,
`Hazard Ratio (HR)` = HR,
`P-value` = P,
`P for Trend` = `P for trend`,
`Confidence Interval` = CI
) %>%
slice(-c(n(), n() - 2))
nafld_results_cleaned %>%
knitr::kable(
digits = 4,
caption = "Cox Proportional Hazards Results for NAFLD Outcome",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Model | Exposure Category | Hazard Ratio (HR) | Confidence Interval | P-value | P for Trend |
|---|---|---|---|---|---|
| Model 1 | expo_lmr_cat2 | 1.068 | 1.022, 1.115 | 0.0033 | 0.0033 |
| Model 1 | expo_lmr_cat3 | 1.105 | 1.049, 1.164 | 0.0002 | NA |
| Model 2 | expo_lmr_cat2 | 1.133 | 1.059, 1.213 | 0.0003 | 0.0003 |
| Model 2 | expo_lmr_cat3 | 1.122 | 1.033, 1.22 | 0.0065 | NA |
| Model 1 | expo_sii_cat2 | 0.824 | 0.789, 0.861 | 0.0000 | 0.0000 |
| Model 1 | expo_sii_cat3 | 0.744 | 0.708, 0.782 | 0.0000 | NA |
| Model 2 | expo_sii_cat2 | 0.851 | 0.795, 0.91 | 0.0000 | 0.0000 |
| Model 2 | expo_sii_cat3 | 0.712 | 0.658, 0.769 | 0.0000 | NA |
| Model 1 | expo_npar_cat2 | 0.873 | 0.835, 0.913 | 0.0000 | 0.0000 |
| Model 1 | expo_npar_cat3 | 0.810 | 0.77, 0.851 | 0.0000 | NA |
| Model 2 | expo_npar_cat2 | 0.864 | 0.806, 0.926 | 0.0000 | 0.0000 |
| Model 2 | expo_npar_cat3 | 0.766 | 0.708, 0.829 | 0.0000 | NA |
| Model 1 | expo_nps_cat2 | 0.840 | 0.795, 0.889 | 0.0000 | 0.0000 |
| Model 2 | expo_nps_cat2 | 0.819 | 0.751, 0.893 | 0.0000 | 0.0000 |
This table presents the results of Cox proportional hazards models
evaluating the association between various baseline exposure categories
and the risk of developing non-alcoholic fatty liver disease. Each
exposure variable (expo_lmr, expo_sii,
expo_npar, expo_nps) is divided into three
categories, with Category 1 (not shown) serving as the reference group
(HR = 1.00). The hazard ratios (HRs), confidence intervals (CIs),
p-values, and p for trend values are provided for two models: Model 1,
adjusted for age and sex, and Model 2, adjusted for additional
covariates including socioeconomic and lifestyle factors.
For expo_lmr, higher exposure categories (cat2 and cat3)
consistently show increased HRs across both models, with slightly higher
HRs in Model 2, indicating a stronger association with NAFLD risk after
full adjustment. In contrast, for expo_sii,
expo_npar, and expo_nps, higher categories
exhibit HRs less than 1 in both models, reflecting their protective
effects against NAFLD. Notably, the protective effect strengthens
slightly in Model 2 for expo_sii and
expo_npar, suggesting that adjusting for additional factors
further clarifies their relationship with NAFLD risk. Significant p for
trend values across most exposures reinforce robust dose-response
relationships, highlighting the importance of these markers in
predicting NAFLD.
Results for Cirrhosis Outcome
cirrhosis_results <- read_csv("./csv/Main_cox_cirrhosis.csv")
cirrhosis_results_cleaned <- cirrhosis_results %>%
select(-1, -lower_conf, -upper_conf) %>%
rename(
Model = Model,
`Exposure Category` = Category,
`Hazard Ratio (HR)` = HR,
`P-value` = P,
`P for Trend` = `P for trend`,
`Confidence Interval` = CI
) %>%
slice(-c(n(), n() - 2))
cirrhosis_results_cleaned %>%
knitr::kable(
digits = 4,
caption = "Cox Proportional Hazards Results for Cirrhosis Outcome",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Model | Exposure Category | Hazard Ratio (HR) | Confidence Interval | P-value | P for Trend |
|---|---|---|---|---|---|
| Model 1 | expo_lmr_cat2 | 0.884 | 0.796, 0.981 | 0.0209 | 0.0209 |
| Model 1 | expo_lmr_cat3 | 0.859 | 0.751, 0.982 | 0.0261 | NA |
| Model 2 | expo_lmr_cat2 | 0.964 | 0.815, 1.14 | 0.6682 | 0.6682 |
| Model 2 | expo_lmr_cat3 | 0.829 | 0.665, 1.033 | 0.0955 | NA |
| Model 1 | expo_sii_cat2 | 0.518 | 0.466, 0.574 | 0.0000 | 0.0000 |
| Model 1 | expo_sii_cat3 | 0.451 | 0.399, 0.509 | 0.0000 | NA |
| Model 2 | expo_sii_cat2 | 0.527 | 0.447, 0.622 | 0.0000 | 0.0000 |
| Model 2 | expo_sii_cat3 | 0.387 | 0.316, 0.474 | 0.0000 | NA |
| Model 1 | expo_npar_cat2 | 0.791 | 0.705, 0.887 | 0.0001 | 0.0001 |
| Model 1 | expo_npar_cat3 | 0.795 | 0.702, 0.9 | 0.0003 | NA |
| Model 2 | expo_npar_cat2 | 0.746 | 0.619, 0.899 | 0.0020 | 0.0020 |
| Model 2 | expo_npar_cat3 | 0.747 | 0.611, 0.915 | 0.0048 | NA |
| Model 1 | expo_nps_cat2 | 0.778 | 0.676, 0.895 | 0.0004 | 0.0004 |
| Model 2 | expo_nps_cat2 | 0.722 | 0.575, 0.906 | 0.0049 | 0.0049 |
Similarly, this table presents the results of Cox proportional
hazards models evaluating the association between various baseline
exposure categories and the risk of developing cirrhosis. For
expo_lmr, higher categories (cat2 and cat3) show HRs
slightly below 1 in Model 1, indicating a modest protective effect
against cirrhosis, but this effect weakens in Model 2, with HRs
approaching 1 and becoming non-significant in some comparisons. In
contrast, expo_sii demonstrates a strong protective
association in both models, with substantially reduced HRs for cat2 and
cat3, and slightly stronger effects observed in Model 2 after full
adjustment. Similarly, expo_npar and expo_nps
consistently show HRs below 1 in both models, with Model 2 indicating
slightly enhanced protective effects compared to Model 1. The
significant p for trend values, particularly for expo_sii
and expo_nps, highlight robust dose-response relationships,
emphasizing the importance of these markers in mitigating cirrhosis
risk.
Conclusion
Discussion
Our study explored the association of lymphocyte-to-monocyte ratio, systemic immune-inflammation index, neutrophil-percentage-to-albumin ratio, and neutrophil-to-platelet score, as the four key inflammatory markers, with the risk for both non-alcoholic fatty liver disease and cirrhosis. With the inclusion of survival analysis, trend of cumulative hazard, and baseline characteristics in this work, our results can be considered a new comprehension regarding the interaction between systemic inflammation and the progression of liver diseases.
For LMR, higher exposure categories (expo_lmr_cat=3) were associated with consistently increased risks of both NAFLD and cirrhosis. The cumulative hazard plots showed a clear dose-response relationship, with higher LMR levels associated with increased cumulative hazard for both outcomes. It suggests that the usual interpretation of LMR as a marker may indicate not reduced inflammation but the dysregulation of adaptive immunity or the prolonged activation of immune competent cells involved in hepatic inflammation and fibrotic processes. Baseline data also showed that the category of intermediate exposure was represented by more of the population, thus suggesting demographic or clinically modifiable variables that may affect the distribution of LMR and modify its association with liver disease risk.
In contrast, SII and NPAR had a protective association with both NAFLD and cirrhosis. In general, higher categories of exposure- namely expo_sii_cat = 3 and expo_npar_cat = 3-were associated with lower cumulative hazard and hazard ratios below 1 across the Cox proportional hazards models, even after adjusting for covariates. These results suggest that higher levels of SII and NPAR reflect compensatory immune mechanisms that attenuate liver injury and are opposite of their established roles as markers of systemic inflammation in other disease contexts.
Indeed, the distribution of baseline characteristics was relatively even across both SII and NPAR categories, reinforcing the stability of these associations and indicating that low levels of these markers reflect a state of increased vulnerability to metabolic stress and immune dysfunction.
In the case of NPS, a similar pattern for protection was observed for cirrhosis, where the higher categories of exposure showed reduced cumulative hazard. However, the baseline data indicated a large population imbalance, where there was an unusually high number of individuals in the low category of exposure (expo_nps_cat=1). This may be one partial explanation for the strength of the association found and points to one of the many reasons accounting for population heterogeneity in survival analyses is important.
In summary, these findings reveal the multifactorial, context-dependent function of systemic inflammation in liver disease. On one hand, inflammation has been an identified driver for NAFLD and cirrhosis; however, the protective associations that SII and NPAR point out clearly indicate the complex nonlinear relationships or threshold effects which stand in the way of any simple-minded interpretation of these markers. Furthermore, the dose-response relationship of significant p-value for trend, observed across all markers, adds to the probability of their predictive utility for the stratification of risk against liver diseases. The integration of the baseline characteristics provides a further clear suggestion as to how demographic and clinical factors can influence such associations. In fact, age, sex, socioeconomic status, and modifiable lifestyle factors might interactively relate with systemic inflammation at the determination of an individual’s risk profile. Mechanistic studies should be conducted in the future to unravel the dual role of inflammatory markers in promoting and mitigating liver disease, exploring how these markers can be integrated into personalized predictive models for improving early detection and targeted interventions in NAFLD and cirrhosis.
Limitations
Although our study illuminates the role of inflammatory markers in predicting the risks of NAFLD and cirrhosis, there are some limitations that warrant mention. The first one would be that this is an observational study in which no causal inferences could be drawn. Moreover, detected associations of LMR, SII, NPAR, and NPS with liver disease outcomes may have been subjected to residual confounding. Although the multivariate models were adjusted for age, sex, socioeconomic status, and lifestyle factors, the influence of unmeasured factors, including genetic predisposition, dietary patterns, and comorbid conditions, may not be entirely eliminated.
Second, the biased distribution of participants in some categories of exposure, especially for NPS, may have biased the results. The excessively large population in the low exposure category of expo_nps_cat = 1 may have inflated the protective association observed for higher categories of exposure. Likewise, subtle differences in baseline characteristics may have influenced the risk estimates across groups, despite the adjustment.
Third, the study was based on single baseline measurements of inflammatory markers, which may not fully reflect the dynamic changes in systemic inflammation. Chronic states of inflammation, fluctuating levels of immune markers, or interventions during follow-up could alter the associations found and therefore misclassify the levels of exposure.
Last but not least, the findings hint at important biologies being involved in the role of inflammatory pathways in liver disease; no such mechanistic data is available to support the interpretation for truly causative pathways. Experimental and longitudinal studies are required that can replicate findings and further dissect whether the protective associations observed were due to a true compensatory immune path or mediated by something.
Given the limited nature of these, it increases the reliability of observed associations where consistency in results arises both from cumulative hazard plots and Cox proportional hazards models. Thus, these markers hold the potential for utility in the stratification of liver disease risk. Research in the future would also be needed to account for these limitations by way of the incorporation of repeated measures of inflammatory markers, exploration of confounding factors, and utilization of mechanistic studies in ways that would enhance this knowledge about the relationships.
Appendix
Baseline Characteristics by LMR Exposure Categories
Baseline_expo_lmr_cat_results <- read_csv("./csv/Baseline_expo_lmr_cat.csv")
Baseline_expo_lmr_cat_cleaned <- Baseline_expo_lmr_cat_results %>%
select(-test) %>%
rename(
` ` = ...1,
Level1 = `1`,
Level2 = `2`,
Level3 = `3`
)
knitr::kable(
Baseline_expo_lmr_cat_cleaned,
digits = 2,
caption = "Baseline Characteristics by LMR Exposure Categories",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Overall | Level1 | Level2 | Level3 | p | |
|---|---|---|---|---|---|
| n | 413146 | 105227 | 204634 | 103279 | NA |
| sex = 2 (%) | 190849 (46.2) | 69149 (65.7) | 93068 (45.5) | 28629 (27.7) | <0.001 |
| age (mean (SD)) | 56.54 (8.09) | 57.70 (8.07) | 56.32 (8.10) | 55.79 (7.96) | <0.001 |
| income (%) | NA | NA | NA | NA | <0.001 |
| 1 | 40844 (9.9) | 9912 (9.4) | 20134 (9.9) | 10798 (10.5) | NA |
| 2 | 17370 (4.2) | 3874 (3.7) | 8290 (4.1) | 5206 (5.1) | NA |
| 3 | 79972 (19.4) | 21680 (20.7) | 37894 (18.6) | 20398 (19.8) | NA |
| 4 | 89874 (21.8) | 23863 (22.8) | 44107 (21.6) | 21902 (21.3) | NA |
| 5 | 92455 (22.4) | 23555 (22.5) | 46634 (22.8) | 22263 (21.6) | NA |
| 6 | 72283 (17.5) | 17494 (16.7) | 37205 (18.2) | 17583 (17.1) | NA |
| 7 | 19249 (4.7) | 4513 (4.3) | 9872 (4.8) | 4864 (4.7) | NA |
| townsend (mean (SD)) | -1.33 (3.08) | -1.39 (3.04) | -1.39 (3.04) | -1.14 (3.18) | <0.001 |
| total_met (mean (SD)) | 1619.08 (2015.70) | 1645.29 (2084.82) | 1625.33 (2008.34) | 1578.85 (1955.34) | <0.001 |
| diet_quality (mean (SD)) | 3.29 (1.57) | 3.12 (1.57) | 3.31 (1.57) | 3.42 (1.56) | <0.001 |
| sleep_hour (mean (SD)) | 7.15 (1.10) | 7.17 (1.12) | 7.15 (1.09) | 7.14 (1.13) | <0.001 |
| smoke_cat (%) | NA | NA | NA | NA | <0.001 |
| 1 | 32255 (7.8) | 6743 (6.4) | 15030 (7.3) | 10481 (10.1) | NA |
| 2 | 11334 (2.7) | 2843 (2.7) | 5572 (2.7) | 2919 (2.8) | NA |
| 3 | 96062 (23.3) | 27859 (26.5) | 47078 (23.0) | 21122 (20.5) | NA |
| 4 | 47431 (11.5) | 12141 (11.5) | 23827 (11.6) | 11463 (11.1) | NA |
| 5 | 60211 (14.6) | 15267 (14.5) | 30436 (14.9) | 14507 (14.0) | NA |
| 6 | 165853 (40.1) | 40374 (38.4) | 82691 (40.4) | 42787 (41.4) | NA |
| total_alcohol (mean (SD)) | 12.55 (15.17) | 15.16 (17.37) | 12.50 (14.77) | 9.98 (12.92) | <0.001 |
| non_hdl (mean (SD)) | 359.49 (78.74) | 345.15 (77.61) | 361.10 (77.88) | 371.09 (79.37) | <0.001 |
| tg (mean (SD)) | 92.17 (22.17) | 92.24 (22.59) | 91.90 (21.60) | 92.66 (22.85) | <0.001 |
| bp_cat (mean (SD)) | 49.88 (28.78) | 47.57 (28.26) | 50.05 (28.71) | 51.91 (29.25) | <0.001 |
Baseline Characteristics by SII Exposure Categories
Baseline_expo_sii_cat_results <- read_csv("./csv/Baseline_expo_sii_cat.csv")
Baseline_expo_sii_cat_cleaned <- Baseline_expo_sii_cat_results %>%
select(-test) %>%
rename(
` ` = ...1,
Level1 = `1`,
Level2 = `2`,
Level3 = `3`
)
knitr::kable(
Baseline_expo_sii_cat_cleaned,
digits = 2,
caption = "Baseline Characteristics by SII Exposure Categories",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Overall | Level1 | Level2 | Level3 | p | |
|---|---|---|---|---|---|
| n | 413146 | 103286 | 206571 | 103286 | NA |
| sex = 2 (%) | 190849 (46.2) | 50744 (49.1) | 93870 (45.4) | 46234 (44.8) | <0.001 |
| age (mean (SD)) | 56.54 (8.09) | 56.70 (7.93) | 56.53 (8.04) | 56.39 (8.34) | <0.001 |
| income (%) | NA | NA | NA | NA | <0.001 |
| 1 | 40844 (9.9) | 10185 (9.9) | 20481 (9.9) | 10178 (9.9) | NA |
| 2 | 17370 (4.2) | 4265 (4.1) | 8296 (4.0) | 4809 (4.7) | NA |
| 3 | 79972 (19.4) | 18085 (17.6) | 38964 (18.9) | 22923 (22.3) | NA |
| 4 | 89874 (21.8) | 22149 (21.5) | 44783 (21.7) | 22941 (22.3) | NA |
| 5 | 92455 (22.4) | 23315 (22.6) | 47003 (22.8) | 22135 (21.5) | NA |
| 6 | 72283 (17.5) | 19303 (18.7) | 36803 (17.9) | 16177 (15.7) | NA |
| 7 | 19249 (4.7) | 5716 (5.5) | 9715 (4.7) | 3818 (3.7) | NA |
| townsend (mean (SD)) | -1.33 (3.08) | -1.30 (3.11) | -1.40 (3.03) | -1.21 (3.13) | <0.001 |
| total_met (mean (SD)) | 1619.08 (2015.70) | 1688.28 (2026.41) | 1612.54 (2009.32) | 1560.73 (2015.54) | <0.001 |
| diet_quality (mean (SD)) | 3.29 (1.57) | 3.42 (1.57) | 3.29 (1.56) | 3.14 (1.57) | <0.001 |
| sleep_hour (mean (SD)) | 7.15 (1.10) | 7.14 (1.08) | 7.16 (1.09) | 7.16 (1.15) | 0.001 |
| smoke_cat (%) | NA | NA | NA | NA | <0.001 |
| 1 | 32255 (7.8) | 6875 (6.7) | 15757 (7.6) | 9622 (9.3) | NA |
| 2 | 11334 (2.7) | 3029 (2.9) | 5568 (2.7) | 2737 (2.6) | NA |
| 3 | 96062 (23.3) | 24195 (23.4) | 47994 (23.2) | 23871 (23.1) | NA |
| 4 | 47431 (11.5) | 12113 (11.7) | 23927 (11.6) | 11391 (11.0) | NA |
| 5 | 60211 (14.6) | 15265 (14.8) | 30389 (14.7) | 14557 (14.1) | NA |
| 6 | 165853 (40.1) | 41809 (40.5) | 82936 (40.1) | 41108 (39.8) | NA |
| total_alcohol (mean (SD)) | 12.55 (15.17) | 12.58 (14.76) | 12.56 (15.01) | 12.49 (15.85) | 0.349 |
| non_hdl (mean (SD)) | 359.49 (78.74) | 358.75 (79.08) | 361.75 (78.52) | 355.70 (78.68) | <0.001 |
| tg (mean (SD)) | 92.17 (22.17) | 91.58 (21.20) | 91.87 (21.41) | 93.38 (24.47) | <0.001 |
| bp_cat (mean (SD)) | 49.88 (28.78) | 51.13 (28.68) | 50.01 (28.76) | 48.38 (28.83) | <0.001 |
Baseline Characteristics by NPAR Exposure Categories
Baseline_expo_npar_cat_results <- read_csv("./csv/Baseline_expo_npar_cat.csv")
Baseline_expo_npar_cat_cleaned <- Baseline_expo_npar_cat_results %>%
select(-test) %>%
rename(
` ` = ...1,
Level1 = `1`,
Level2 = `2`,
Level3 = `3`
)
knitr::kable(
Baseline_expo_npar_cat_cleaned,
digits = 2,
caption = "Baseline Characteristics by NPAR Exposure Categories",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Overall | Level1 | Level2 | Level3 | p | |
|---|---|---|---|---|---|
| n | 413146 | 103287 | 206574 | 103285 | NA |
| sex = 2 (%) | 190849 (46.2) | 48556 (47.0) | 95248 (46.1) | 47045 (45.5) | <0.001 |
| age (mean (SD)) | 56.54 (8.09) | 55.91 (7.89) | 56.54 (8.05) | 57.16 (8.31) | <0.001 |
| income (%) | NA | NA | NA | NA | <0.001 |
| 1 | 40844 (9.9) | 10029 (9.7) | 20412 (9.9) | 10403 (10.1) | NA |
| 2 | 17370 (4.2) | 4084 (4.0) | 8458 (4.1) | 4828 (4.7) | NA |
| 3 | 79972 (19.4) | 16934 (16.4) | 39005 (18.9) | 24033 (23.3) | NA |
| 4 | 89874 (21.8) | 21489 (20.9) | 44821 (21.8) | 23564 (22.9) | NA |
| 5 | 92455 (22.4) | 23997 (23.3) | 46929 (22.8) | 21529 (20.9) | NA |
| 6 | 72283 (17.5) | 20577 (20.0) | 36749 (17.8) | 14957 (14.5) | NA |
| 7 | 19249 (4.7) | 5948 (5.8) | 9688 (4.7) | 3613 (3.5) | NA |
| townsend (mean (SD)) | -1.33 (3.08) | -1.34 (3.10) | -1.40 (3.03) | -1.17 (3.14) | <0.001 |
| total_met (mean (SD)) | 1619.08 (2015.70) | 1671.02 (2018.22) | 1617.55 (2012.63) | 1568.39 (2018.03) | <0.001 |
| diet_quality (mean (SD)) | 3.29 (1.57) | 3.41 (1.57) | 3.29 (1.57) | 3.16 (1.57) | <0.001 |
| sleep_hour (mean (SD)) | 7.15 (1.10) | 7.13 (1.08) | 7.15 (1.09) | 7.18 (1.16) | <0.001 |
| smoke_cat (%) | NA | NA | NA | NA | <0.001 |
| 1 | 32255 (7.8) | 6618 (6.4) | 15628 (7.6) | 10009 (9.7) | NA |
| 2 | 11334 (2.7) | 3051 (3.0) | 5572 (2.7) | 2711 (2.6) | NA |
| 3 | 96062 (23.3) | 24055 (23.3) | 48085 (23.3) | 23922 (23.2) | NA |
| 4 | 47431 (11.5) | 12149 (11.8) | 23838 (11.5) | 11444 (11.1) | NA |
| 5 | 60211 (14.6) | 15418 (14.9) | 30402 (14.7) | 14391 (13.9) | NA |
| 6 | 165853 (40.1) | 41996 (40.7) | 83049 (40.2) | 40808 (39.5) | NA |
| total_alcohol (mean (SD)) | 12.55 (15.17) | 12.98 (15.08) | 12.63 (15.06) | 11.95 (15.44) | <0.001 |
| non_hdl (mean (SD)) | 359.49 (78.74) | 368.07 (79.61) | 361.33 (78.16) | 347.34 (77.58) | <0.001 |
| tg (mean (SD)) | 92.17 (22.17) | 90.77 (18.16) | 91.81 (21.11) | 94.30 (27.18) | <0.001 |
| bp_cat (mean (SD)) | 49.88 (28.78) | 49.89 (28.39) | 49.86 (28.70) | 49.93 (29.30) | 0.806 |
Baseline Characteristics by NPS Exposure Categories
Baseline_expo_nps_cat_results <- read_csv("./csv/Baseline_expo_nps_cat.csv")
Baseline_expo_nps_cat_cleaned <- Baseline_expo_nps_cat_results %>%
select(-test) %>%
rename(
` ` = ...1,
Level1 = `1`,
Level2 = `2`
)
knitr::kable(
Baseline_expo_nps_cat_cleaned,
digits = 2,
caption = "Baseline Characteristics by NPS Exposure Categories",
align = "c"
) %>%
kable_styling(full_width = FALSE, position = "center")| Overall | Level1 | Level2 | p | |
|---|---|---|---|---|
| n | 413146 | 365846 | 47294 | NA |
| sex = 2 (%) | 190849 (46.2) | 164549 (45.0) | 26297 (55.6) | <0.001 |
| age (mean (SD)) | 56.54 (8.09) | 56.45 (8.06) | 57.22 (8.26) | <0.001 |
| income (%) | NA | NA | NA | <0.001 |
| 1 | 40844 (9.9) | 36184 (9.9) | 4660 (9.9) | NA |
| 2 | 17370 (4.2) | 15368 (4.2) | 2002 (4.2) | NA |
| 3 | 79972 (19.4) | 69618 (19.1) | 10354 (22.0) | NA |
| 4 | 89874 (21.8) | 79112 (21.7) | 10760 (22.8) | NA |
| 5 | 92455 (22.4) | 82188 (22.5) | 10264 (21.8) | NA |
| 6 | 72283 (17.5) | 65052 (17.8) | 7230 (15.3) | NA |
| 7 | 19249 (4.7) | 17374 (4.8) | 1875 (4.0) | NA |
| townsend (mean (SD)) | -1.33 (3.08) | -1.33 (3.08) | -1.31 (3.08) | 0.219 |
| total_met (mean (SD)) | 1619.08 (2015.70) | 1620.03 (2009.37) | 1611.68 (2064.33) | 0.457 |
| diet_quality (mean (SD)) | 3.29 (1.57) | 3.31 (1.57) | 3.14 (1.57) | <0.001 |
| sleep_hour (mean (SD)) | 7.15 (1.10) | 7.15 (1.10) | 7.17 (1.14) | 0.005 |
| smoke_cat (%) | NA | NA | NA | <0.001 |
| 1 | 32255 (7.8) | 29492 (8.1) | 2762 (5.8) | NA |
| 2 | 11334 (2.7) | 10183 (2.8) | 1151 (2.4) | NA |
| 3 | 96062 (23.3) | 84650 (23.1) | 11409 (24.1) | NA |
| 4 | 47431 (11.5) | 41980 (11.5) | 5451 (11.5) | NA |
| 5 | 60211 (14.6) | 53024 (14.5) | 7186 (15.2) | NA |
| 6 | 165853 (40.1) | 146517 (40.0) | 19335 (40.9) | NA |
| total_alcohol (mean (SD)) | 12.55 (15.17) | 12.42 (14.99) | 13.52 (16.42) | <0.001 |
| non_hdl (mean (SD)) | 359.49 (78.74) | 360.84 (78.79) | 349.07 (77.57) | <0.001 |
| tg (mean (SD)) | 92.17 (22.17) | 91.95 (21.76) | 93.91 (25.10) | <0.001 |
| bp_cat (mean (SD)) | 49.88 (28.78) | 50.14 (28.77) | 47.86 (28.74) | <0.001 |
The baseline characteristics tables present a comprehensive summary of the study population stratified by four key exposure categories: Lymphocyte-to-Monocyte Ratio (LMR), Systemic Immune-Inflammation Index (SII), Neutrophil-to-Albumin Ratio (NPAR), and Naples Prognostic Score (NPS). These tables outline demographic variables (e.g., age, sex), socioeconomic factors (e.g., income, Townsend index), lifestyle behaviors (e.g., smoking, alcohol intake), and clinical markers (e.g., triglycerides, diet quality), while highlighting statistically significant differences across exposure levels for most variables. These differences underscore the potential influence of these covariates on the outcomes, necessitating their adjustment in subsequent analyses, such as Cox proportional hazards modeling, to mitigate confounding and ensure robust evaluation of exposure-outcome relationships.