Exploratory Data Analysis
ukb_data <- read_dta("./external_data/data_prepared.dta")To understand the data structure and for further EDA, we initially examined the associations between key demographic and lifestyle variables, such as age, sex, alcohol consumption, blood pressure, income, diet quality, and sleep duration. This step aimed to identify potential patterns and relationships between participants’ characteristics and overall health outcomes, laying the groundwork for subsequent analyses.
To facilitate the initial understanding of inflammatory indices and for further survival analysis, we explored the relationships between blood parameters (e.g., lymphocyte-to-monocyte ratio, systemic immune-inflammation index) and exposure variables with disease outcomes like NAFLD and cirrhosis. This step provided insights into the predictive value of these indices and their interactions with other variables, offering a foundation for examining disease progression in later stages of the project.
Age and Sex Distribution
figure_1 <- ukb_data %>%
mutate(age_group = cut(age, breaks = seq(10, 100, by = 10), right = FALSE,
labels = paste(seq(10, 90, by = 10), "-",
seq(19, 99, by = 10), sep = ""))) %>%
ggplot(aes(x = age_group, y = age, fill = as.factor(sex))) +
geom_boxplot(alpha = 0.9) +
theme_minimal() +
labs(x = 'Age Groups',
y = 'Age',
title = 'Age Distribution by Sex and Age Groups',
fill = 'Sex') +
scale_fill_manual(values = c("lightblue", "lightpink"),
labels = c("Female", "Male")) +
theme(legend.position = "bottom")
figure_1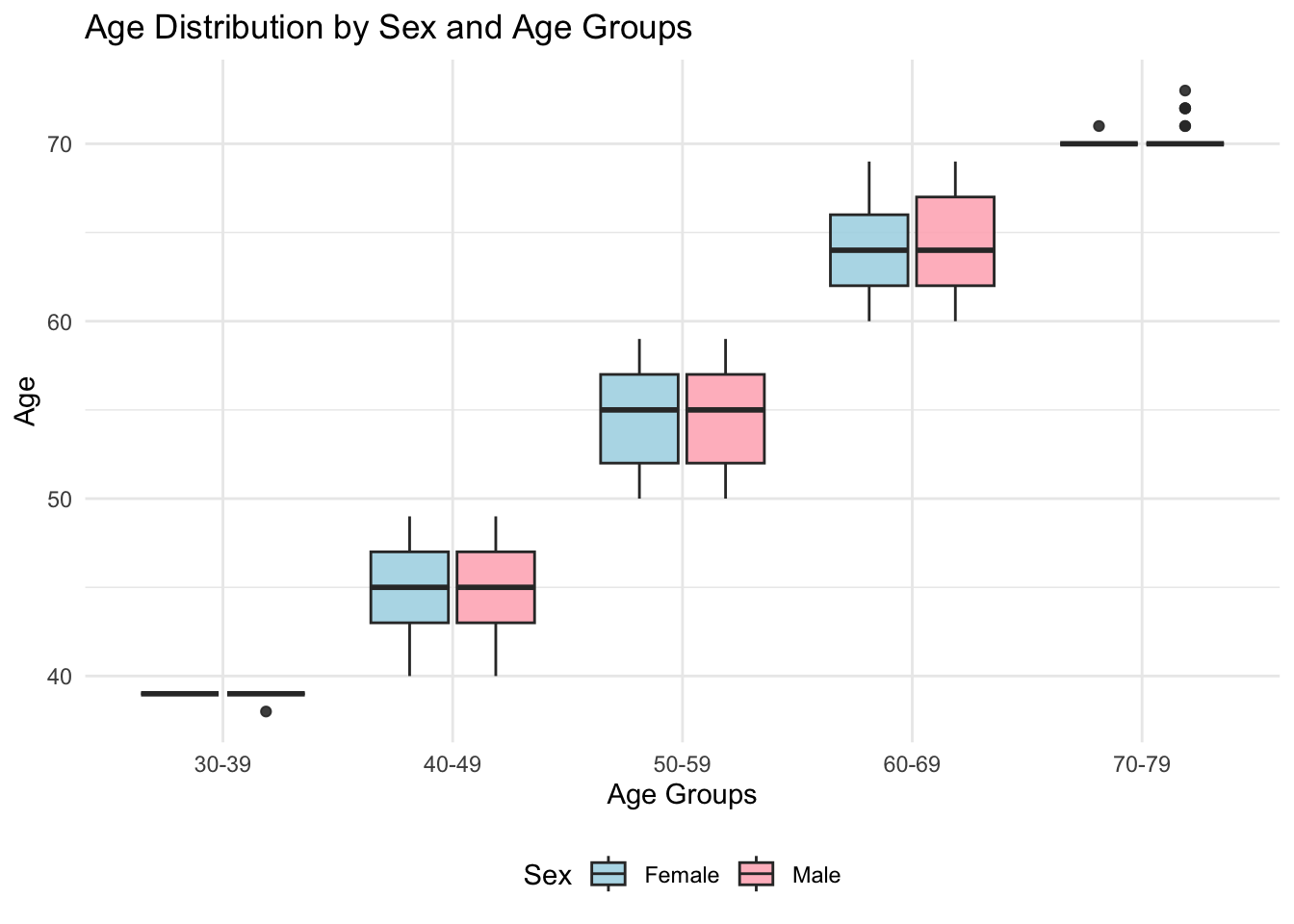
We examined the age distribution of male and female participants across 10-year age groups. In the 30-39 age group, the sample size was relatively small, with median ages and IQRs consistent across sexes. Similar trends were observed in the 40-49 and 50-59 groups. In the 60-69 group, females exhibited slightly higher upper quartiles than males, while the 70-79 group showed broader distributions for females, indicating more variation in ages.
Alcohol Consumption and Disease Incidence
# Data cleaning
disease_heat2 <- ukb_data %>%
filter(!is.na(sex), !is.na(total_alcohol), !is.na(nafld_outcome)) %>%
mutate(
alcohol_group = cut(total_alcohol,
breaks = c(-Inf, 0, 5, 10, 15, Inf),
labels = c("None", "Low", "Medium", "High", "Very High"),
include.lowest = TRUE)
) %>%
group_by(sex, alcohol_group) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
ukb_data <- ukb_data %>%
mutate(
sex = as_factor(sex),
nafld_outcome = as.numeric(as_factor(nafld_outcome)),
total_alcohol = as.numeric(total_alcohol),
income = as.numeric(income)
)
disease_heat2 <- ukb_data %>%
filter(!is.na(sex), !is.na(total_alcohol), !is.na(nafld_outcome)) %>%
mutate(
alcohol_group = cut(total_alcohol,
breaks = c(-Inf, 0, 5, 10, 15, Inf),
labels = c("None", "Low", "Medium", "High", "Very High"),
include.lowest = TRUE)
) %>%
group_by(sex, alcohol_group) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
# Create heatmap
figure_2 <- ggplot(disease_heat2, aes(x = sex, y = alcohol_group, fill = disease_rate)) +
geom_tile() +
scale_fill_gradient(low = "white", high = "red") +
theme_minimal() +
labs(title = "Disease Incidence by Sex and Alcohol Consumption",
x = "Sex",
y = "Alcohol Consumption Level",
fill = "Disease Rate") +
geom_text(aes(label = sprintf("n=%d", n)), color = "black", size = 3)
ggplotly(figure_2)A heatmap analysis revealed an inverse relationship between alcohol consumption and disease incidence. Non-drinkers exhibited the highest disease rates, with males outnumbering females. In contrast, participants with very high alcohol consumption had the lowest disease rates. Across all categories, females demonstrated slightly higher disease rates than males.
Blood Pressure and Age Trends
# Data Cleaning
disease_heat1 <- ukb_data %>%
filter(!is.na(age), !is.na(bp_cat), !is.na(nafld_outcome)) %>%
mutate(
bp_cat = as_factor(bp_cat),
age_group = cut(age,
breaks = c(30, 40, 50, 60, 70, 80),
labels = c("30-40", "40-50", "50-60", "60-70", "70-80"),
include.lowest = TRUE)
) %>%
group_by(age_group, bp_cat) %>%
summarise(
disease_rate = mean(nafld_outcome, na.rm = TRUE),
n = n()
) %>%
ungroup()
# Create heatmap
figure_4 <- ggplot(disease_heat1, aes(x = age_group, y = bp_cat, fill = disease_rate)) +
geom_tile() +
scale_fill_gradient(low = "white", high = "red") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45)) +
labs(title = "Disease Incidence by Age and Blood Pressure Category",
x = "Age Groups",
y = "BP Category",
fill = "Disease Rate") +
geom_text(aes(label = sprintf("n=%d", n)), color = "black", size = 3)
ggplotly(figure_4)We observed that disease incidence increased significantly with higher blood pressure, particularly in individuals aged 60 and above. Younger age groups (30-40) showed low disease incidence even at higher blood pressure levels, while middle-aged and older groups demonstrated steeper increases. The largest sample sizes were concentrated in moderate blood pressure categories (BP=40-60).
Income and Diet Quality
color_palette <- colorRampPalette(c("lightblue", "darkblue"))(length(unique(ukb_data$diet_quality)))
figure_4 <- ukb_data %>%
filter(!is.na(income) & !is.na(diet_quality)) %>%
ggplot(aes(x = as.factor(income), fill = as.factor(diet_quality))) +
geom_bar(position = "stack") +
theme_minimal() +
scale_fill_manual(values = color_palette) +
labs(x = "Income Levels",
y = "Count",
fill = "Diet Quality",
title = "Income vs Diet Quality") +
theme(legend.position = "bottom")
ggplotly(figure_4)Diet quality showed a positive correlation with income levels. Participants in the highest income bracket had the highest proportions of top diet quality, while those in the lowest income bracket exhibited the poorest diet quality. Middle-income groups displayed more balanced distributions.
Sleep Duration Across Age Groups
figure_5 <- ukb_data %>%
filter(!is.na(age) & !is.na(sleep_hour)) %>%
mutate(age_group = cut(age, breaks = seq(10, 100, by = 10), right = FALSE,
labels = paste(seq(10, 90, by = 10), "-",
seq(19, 99, by = 10), sep = ""))) %>%
ggplot(aes(x = age_group, y = sleep_hour, fill = age_group)) +
geom_boxplot() +
theme_minimal() +
labs(x = "Age Groups",
y = "Sleep Hours per Night",
title = "Sleep Hours per Night by Age Groups",
fill = "Age Groups") +
scale_fill_manual(values = c("lightgreen", "lightpink", "lightblue",
"lightcoral", "lightgoldenrod"))
figure_5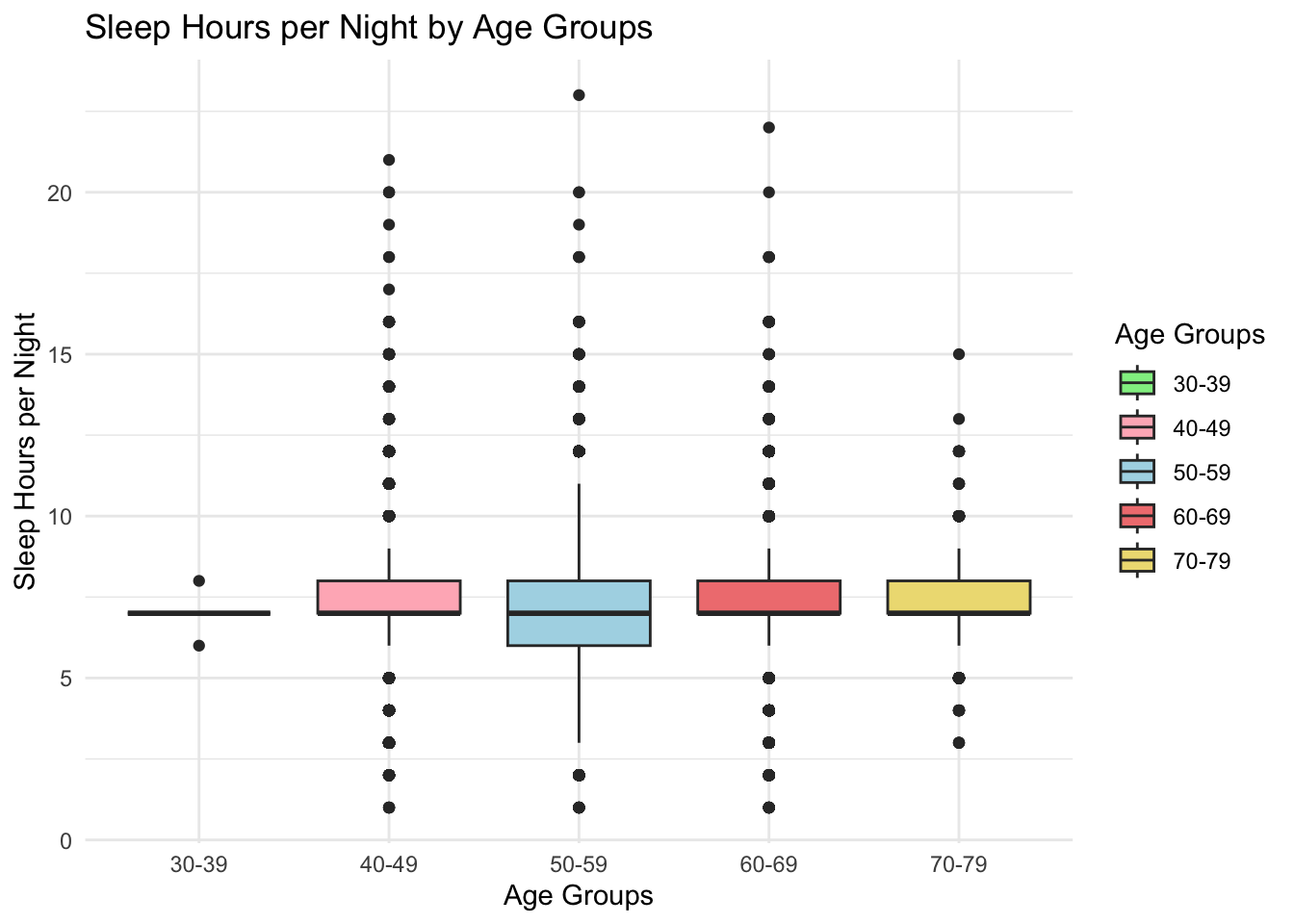
Sleep duration varied slightly by age. Younger participants (30-39) had the shortest median sleep duration and the highest prevalence of short sleep (<6 hours). Older age groups (60-69, 70-79) displayed greater variability, with outliers on both ends of the sleep duration spectrum, suggesting more diverse sleep patterns in older adults.
Exposure variables and disease outcomes
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_lmr)) %>%
summarize(
q25 = quantile(expo_lmr, 0.25),
median = median(expo_lmr),
q75 = quantile(expo_lmr, 0.75))
figure_6 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_lmr)) %>%
mutate(
lower_bound = quantile(expo_lmr, 0.25) - 1.5 * IQR(expo_lmr),
upper_bound = quantile(expo_lmr, 0.75) + 1.5 * IQR(expo_lmr)
) %>%
filter(expo_lmr >= lower_bound & expo_lmr <= upper_bound) %>%
ggplot(aes(x = expo_lmr, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte-to-Monocyte Ratio (LMR)",
y = "Density",
title = "Density Plot of expo_lmr by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_sii)) %>%
summarize(
q25 = quantile(expo_sii, 0.25),
median = median(expo_sii),
q75 = quantile(expo_sii, 0.75)
)
figure_7 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_sii)) %>%
mutate(
lower_bound = quantile(expo_sii, 0.25) - 1.5 * IQR(expo_sii),
upper_bound = quantile(expo_sii, 0.75) + 1.5 * IQR(expo_sii)
) %>%
filter(expo_sii >= lower_bound & expo_sii <= upper_bound) %>%
ggplot(aes(x = expo_sii, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Systemic Immune-Inflammation Index (SII)",
y = "Density",
title = "Density Plot of expo_sii by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_npar)) %>%
summarize(
q25 = quantile(expo_npar, 0.25),
median = median(expo_npar),
q75 = quantile(expo_npar, 0.75)
)
figure_8 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(expo_npar)) %>%
mutate(
lower_bound = quantile(expo_npar, 0.25) - 1.5 * IQR(expo_npar),
upper_bound = quantile(expo_npar, 0.75) + 1.5 * IQR(expo_npar)
) %>%
filter(expo_npar >= lower_bound & expo_npar <= upper_bound) %>%
ggplot(aes(x = expo_npar, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophil-to-Albumin Ratio (NPAR)",
y = "Density",
title = "Density Plot of expo_npar by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
figure_9 <-ukb_data %>%
filter(!is.na(expo_nps)) %>%
group_by(nafld_outcome, expo_nps) %>%
summarize(count = n()) %>%
ungroup() %>%
mutate(total_count = sum(count),
percentage = count / total_count * 100) %>%
ggplot(aes(x = as.factor(expo_nps), y = percentage, fill = as.factor(nafld_outcome))) +
geom_bar(stat = "identity", position = position_dodge(width = 0.9), alpha = 0.8) +
geom_text(aes(label = paste0(round(percentage, 1), "%")),
position = position_dodge(width = 0.9), vjust = 0.75, size = 3) +
scale_fill_manual(values = c("skyblue", "pink"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Naples Prognostic Score (NPS)",
y = "Percentage of Total Population",
title = "Percentage of Total Population by NPS and NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
)
grid.arrange(figure_6, figure_7, figure_8, figure_9, nrow = 2, ncol = 2)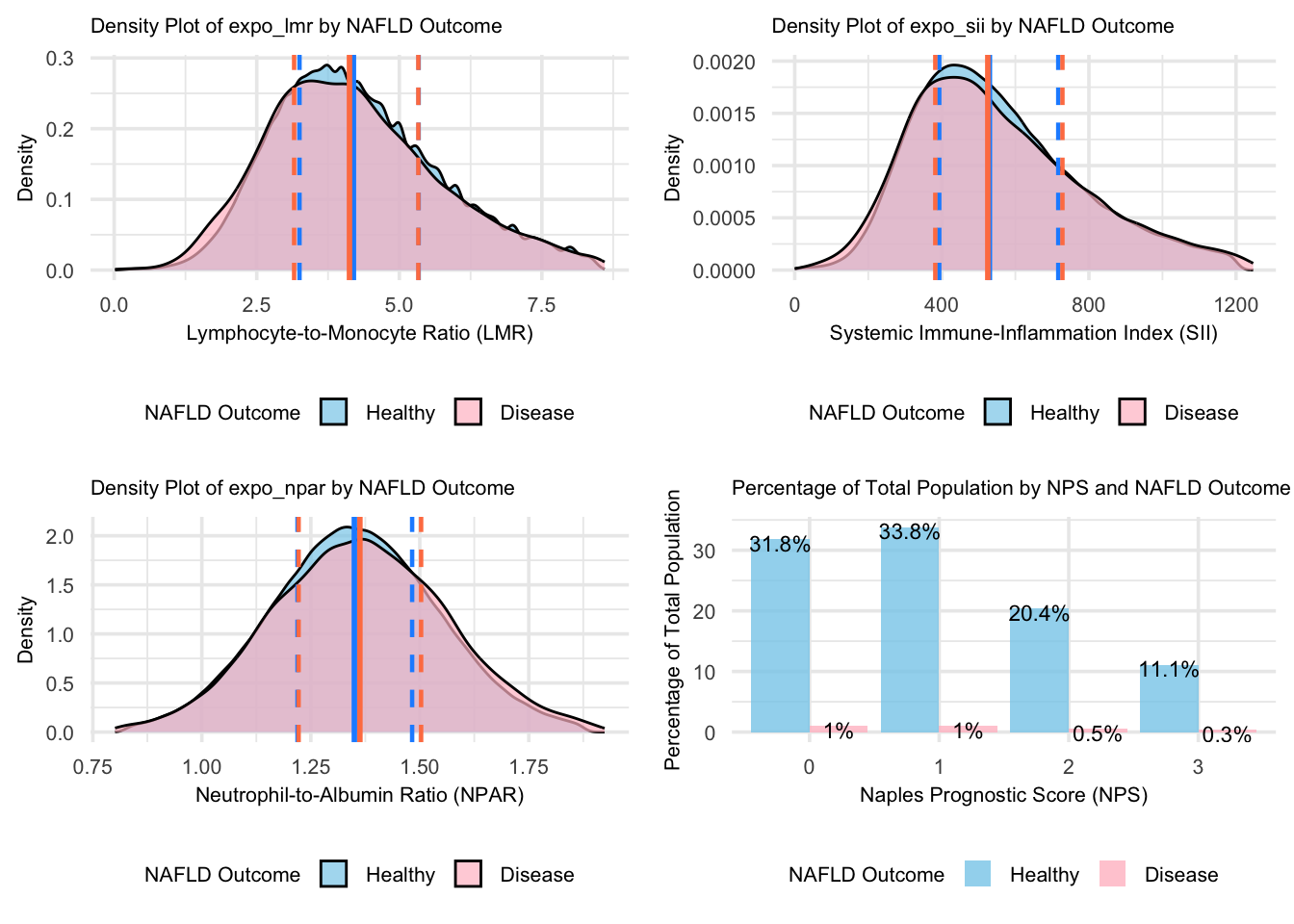
For the continuous biomarkers (LMR, SII, and NPAR), the density plots show consistently lower concentrations of values in the NAFLD group compared to the Healthy group, indicating a complex immune-inflammatory profile in NAFLD pathogenesis. The lower density peaks in LMR reflect more dispersed immune balance patterns, while decreased density in SII suggests more variable inflammatory responses. Similarly, the broader distribution of NPAR in NAFLD may indicate more heterogeneous liver function and neutrophil activity patterns. The vertical lines in each density plot represent the quartiles (Q1=25th percentile, Q2=median, and Q3=75th percentile), providing a clear visualization of the data distribution. The close median values and significant distribution overlap suggest limited discriminative power for these biomarkers alone in distinguishing NAFLD from healthy individuals. The NPS distribution shows higher proportions of healthy individuals across all scores, particularly in lower-risk categories (0-1), highlighting that while NAFLD alters immune-inflammatory profiles, this does not necessarily translate into higher prognostic risks.
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_lmr)) %>%
summarize(
q25 = quantile(expo_lmr, 0.25),
median = median(expo_lmr),
q75 = quantile(expo_lmr, 0.75))
figure_10 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_lmr)) %>%
mutate(
lower_bound = quantile(expo_lmr, 0.25) - 1.5 * IQR(expo_lmr),
upper_bound = quantile(expo_lmr, 0.75) + 1.5 * IQR(expo_lmr)
) %>%
filter(expo_lmr >= lower_bound & expo_lmr <= upper_bound) %>%
ggplot(aes(x = expo_lmr, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte-to-Monocyte Ratio (LMR)",
y = "Density",
title = "Density Plot of expo_lmr by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_sii)) %>%
summarize(
q25 = quantile(expo_sii, 0.25),
median = median(expo_sii),
q75 = quantile(expo_sii, 0.75)
)
figure_11 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_sii)) %>%
mutate(
lower_bound = quantile(expo_sii, 0.25) - 1.5 * IQR(expo_sii),
upper_bound = quantile(expo_sii, 0.75) + 1.5 * IQR(expo_sii)
) %>%
filter(expo_sii >= lower_bound & expo_sii <= upper_bound) %>%
ggplot(aes(x = expo_sii, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Systemic Immune-Inflammation Index (SII)",
y = "Density",
title = "Density Plot of expo_sii by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_npar)) %>%
summarize(
q25 = quantile(expo_npar, 0.25),
median = median(expo_npar),
q75 = quantile(expo_npar, 0.75)
)
figure_12 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(expo_npar)) %>%
mutate(
lower_bound = quantile(expo_npar, 0.25) - 1.5 * IQR(expo_npar),
upper_bound = quantile(expo_npar, 0.75) + 1.5 * IQR(expo_npar)
) %>%
filter(expo_npar >= lower_bound & expo_npar <= upper_bound) %>%
ggplot(aes(x = expo_npar, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophil-to-Albumin Ratio (NPAR)",
y = "Density",
title = "Density Plot of expo_npar by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("dodgerblue", "coral"))
figure_13 <- ukb_data %>%
filter(!is.na(expo_nps)) %>%
group_by(cirrho_outcome, expo_nps) %>%
summarize(count = n()) %>%
ungroup() %>%
mutate(total_count = sum(count),
percentage = count / total_count * 100) %>%
ggplot(aes(x = as.factor(expo_nps), y = percentage, fill = as.factor(cirrho_outcome))) +
geom_bar(stat = "identity", position = position_dodge(width = 0.9), alpha = 0.8) +
geom_text(aes(label = paste0(round(percentage, 1), "%")),
position = position_dodge(width = 0.9), vjust = 0.5, size = 3) +
scale_fill_manual(values = c("skyblue", "pink"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Naples Prognostic Score (NPS)",
y = "Percentage of Total Population",
title = "Percentage of Total Population by NPS and Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
)
grid.arrange(figure_10, figure_11, figure_12, figure_13, nrow = 2, ncol = 2)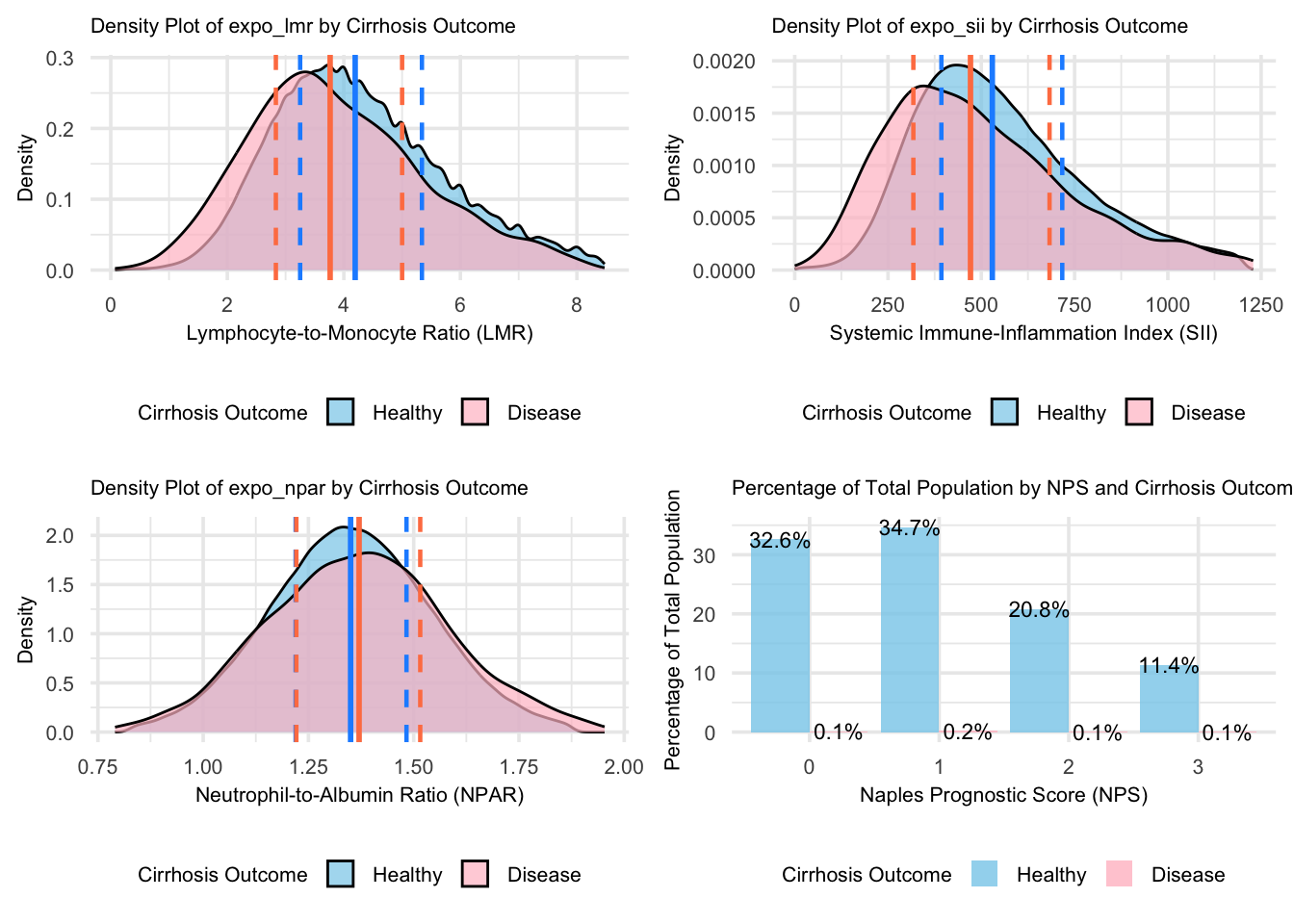
For the Cirrhosis group, the density plots reveal distinct distribution patterns compared to the Healthy group: LMR and SII show lower peak heights in cirrhosis, indicating more dispersed values and suggesting variable immune and inflammatory states, while NPAR shows shifted peak concentrations, reflecting altered liver function patterns. The height and shape of the density curves suggest more concentrated value distributions in healthy individuals for LMR and SII, indicating more consistent immune-inflammatory states in health. Though the overlapping areas between curves indicate some shared value ranges, the different peak heights and positions reflect distinct disease characteristics. The NPS distribution further emphasizes this contrast, with higher density concentrations in healthy individuals across risk categories.
Blood parameters and disease outcomes
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_lympho)) %>%
summarize(
q25 = quantile(b_lympho, 0.25),
median = median(b_lympho),
q75 = quantile(b_lympho, 0.75))
figure_14 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_lympho)) %>%
mutate(
lower_bound = quantile(b_lympho, 0.25) - 1.5 * IQR(b_lympho),
upper_bound = quantile(b_lympho, 0.75) + 1.5 * IQR(b_lympho)
) %>%
filter(b_lympho >= lower_bound & b_lympho <= upper_bound) %>%
ggplot(aes(x = b_lympho, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte",
y = "Density",
title = "Density Plot of b_lympho by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_mono)) %>%
summarize(
q25 = quantile(b_mono, 0.25),
median = median(b_mono),
q75 = quantile(b_mono, 0.75)
)
figure_15 <-ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_mono)) %>%
mutate(
lower_bound = quantile(b_mono, 0.25) - 1.5 * IQR(b_mono),
upper_bound = quantile(b_mono, 0.75) + 1.5 * IQR(b_mono)
) %>%
filter(b_mono >= lower_bound & b_mono <= upper_bound) %>%
ggplot(aes(x = b_mono, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Monocytes",
y = "Density",
title = "Density Plot of b_mono by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_neutro)) %>%
summarize(
q25 = quantile(b_neutro, 0.25),
median = median(b_neutro),
q75 = quantile(b_neutro, 0.75)
)
figure_16 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_neutro)) %>%
mutate(
lower_bound = quantile(b_neutro, 0.25) - 1.5 * IQR(b_neutro),
upper_bound = quantile(b_neutro, 0.75) + 1.5 * IQR(b_neutro)
) %>%
filter(b_neutro >= lower_bound & b_neutro <= upper_bound) %>%
ggplot(aes(x = b_neutro, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophils",
y = "Density",
title = "Density Plot of b_neutro by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_plate)) %>%
summarize(
q25 = quantile(b_plate, 0.25),
median = median(b_plate),
q75 = quantile(b_plate, 0.75)
)
figure_17 <- ukb_data %>%
group_by(nafld_outcome) %>%
filter(!is.na(b_plate)) %>%
mutate(
lower_bound = quantile(b_plate, 0.25) - 1.5 * IQR(b_plate),
upper_bound = quantile(b_plate, 0.75) + 1.5 * IQR(b_plate)
) %>%
filter(b_plate >= lower_bound & b_plate <= upper_bound) %>%
ggplot(aes(x = b_plate, fill = as.factor(nafld_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "NAFLD Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Platelets",
y = "Density",
title = "Density Plot of b_plate by NAFLD Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(nafld_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(nafld_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
grid.arrange(figure_14, figure_15, figure_16, figure_17, nrow = 2, ncol = 2)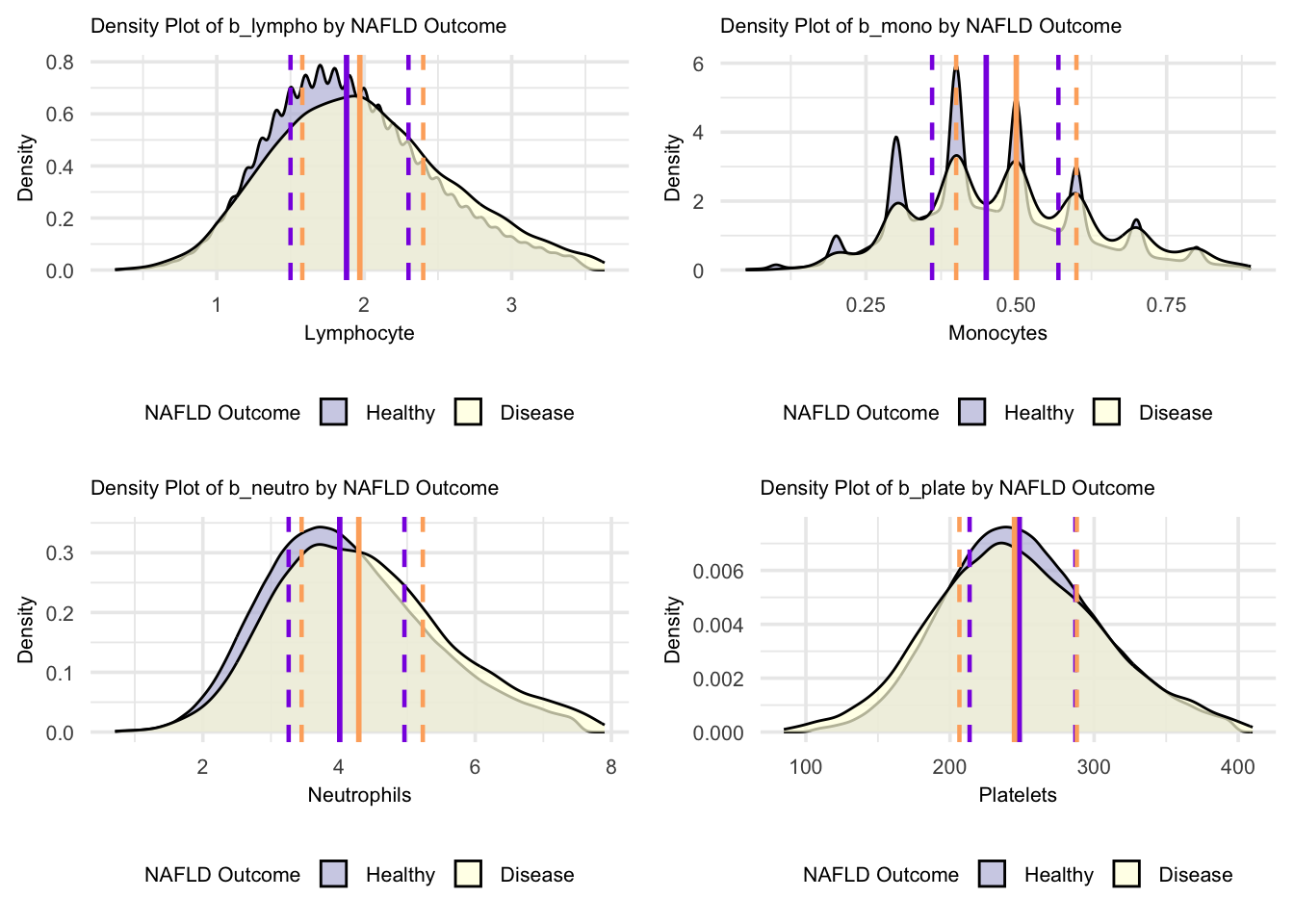
For these blood parameters (lymphocytes, monocytes, neutrophils, and platelets), the density plots reveal distinct distribution patterns. The monocyte plot shows multiple sharp peaks, indicating distinct clustered values in both groups. Lymphocyte and neutrophil distributions show bell-shaped curves with considerable overlap but different peak heights, suggesting concentrated value ranges in both healthy and NAFLD states. Platelet distributions show broader, more spread-out patterns with similar shapes between groups, indicating more variable platelet counts in both populations. These density patterns suggest that while the value ranges overlap, the concentration of measurements differs between healthy and NAFLD states, with each parameter showing unique distribution characteristics.
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_lympho)) %>%
summarize(
q25 = quantile(b_lympho, 0.25),
median = median(b_lympho),
q75 = quantile(b_lympho, 0.75))
figure_18 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_lympho)) %>%
mutate(
lower_bound = quantile(b_lympho, 0.25) - 1.5 * IQR(b_lympho),
upper_bound = quantile(b_lympho, 0.75) + 1.5 * IQR(b_lympho)
) %>%
filter(b_lympho >= lower_bound & b_lympho <= upper_bound) %>%
ggplot(aes(x = b_lympho, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Lymphocyte",
y = "Density",
title = "Density Plot of b_lympho by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_mono)) %>%
summarize(
q25 = quantile(b_mono, 0.25),
median = median(b_mono),
q75 = quantile(b_mono, 0.75)
)
figure_19 <-ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_mono)) %>%
mutate(
lower_bound = quantile(b_mono, 0.25) - 1.5 * IQR(b_mono),
upper_bound = quantile(b_mono, 0.75) + 1.5 * IQR(b_mono)
) %>%
filter(b_mono >= lower_bound & b_mono <= upper_bound) %>%
ggplot(aes(x = b_mono, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Monocytes",
y = "Density",
title = "Density Plot of b_mono by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_neutro)) %>%
summarize(
q25 = quantile(b_neutro, 0.25),
median = median(b_neutro),
q75 = quantile(b_neutro, 0.75)
)
figure_20 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_neutro)) %>%
mutate(
lower_bound = quantile(b_neutro, 0.25) - 1.5 * IQR(b_neutro),
upper_bound = quantile(b_neutro, 0.75) + 1.5 * IQR(b_neutro)
) %>%
filter(b_neutro >= lower_bound & b_neutro <= upper_bound) %>%
ggplot(aes(x = b_neutro, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Neutrophils",
y = "Density",
title = "Density Plot of b_neutro by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
quantiles <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_plate)) %>%
summarize(
q25 = quantile(b_plate, 0.25),
median = median(b_plate),
q75 = quantile(b_plate, 0.75)
)
figure_21 <- ukb_data %>%
group_by(cirrho_outcome) %>%
filter(!is.na(b_plate)) %>%
mutate(
lower_bound = quantile(b_plate, 0.25) - 1.5 * IQR(b_plate),
upper_bound = quantile(b_plate, 0.75) + 1.5 * IQR(b_plate)
) %>%
filter(b_plate >= lower_bound & b_plate <= upper_bound) %>%
ggplot(aes(x = b_plate, fill = as.factor(cirrho_outcome))) +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#BCBDDC", "lightyellow"), name = "Cirrhosis Outcome",
labels = c("Healthy", "Disease")) +
labs(
x = "Platelets",
y = "Density",
title = "Density Plot of b_plate by Cirrhosis Outcome"
) +
theme_minimal(base_size = 14) +
theme(
legend.position = "bottom",
plot.title = element_text(size = 8),
axis.title.x = element_text(size = 8),
axis.title.y = element_text(size = 8),
axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 8),
legend.title = element_text(size = 8),
legend.text = element_text(size = 8),
legend.key.size = unit(0.4, "cm")
) +
geom_vline(data = quantiles, aes(xintercept = q25,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = median,
color = as.factor(cirrho_outcome)),
linetype = "solid", size = 1, show.legend = FALSE) +
geom_vline(data = quantiles, aes(xintercept = q75,
color = as.factor(cirrho_outcome)),
linetype = "dashed", size = 0.8, show.legend = FALSE) +
scale_color_manual(values = c("#8A2BE2", "#FDAE6B"))
grid.arrange(figure_18, figure_19, figure_20, figure_21, nrow = 2, ncol = 2)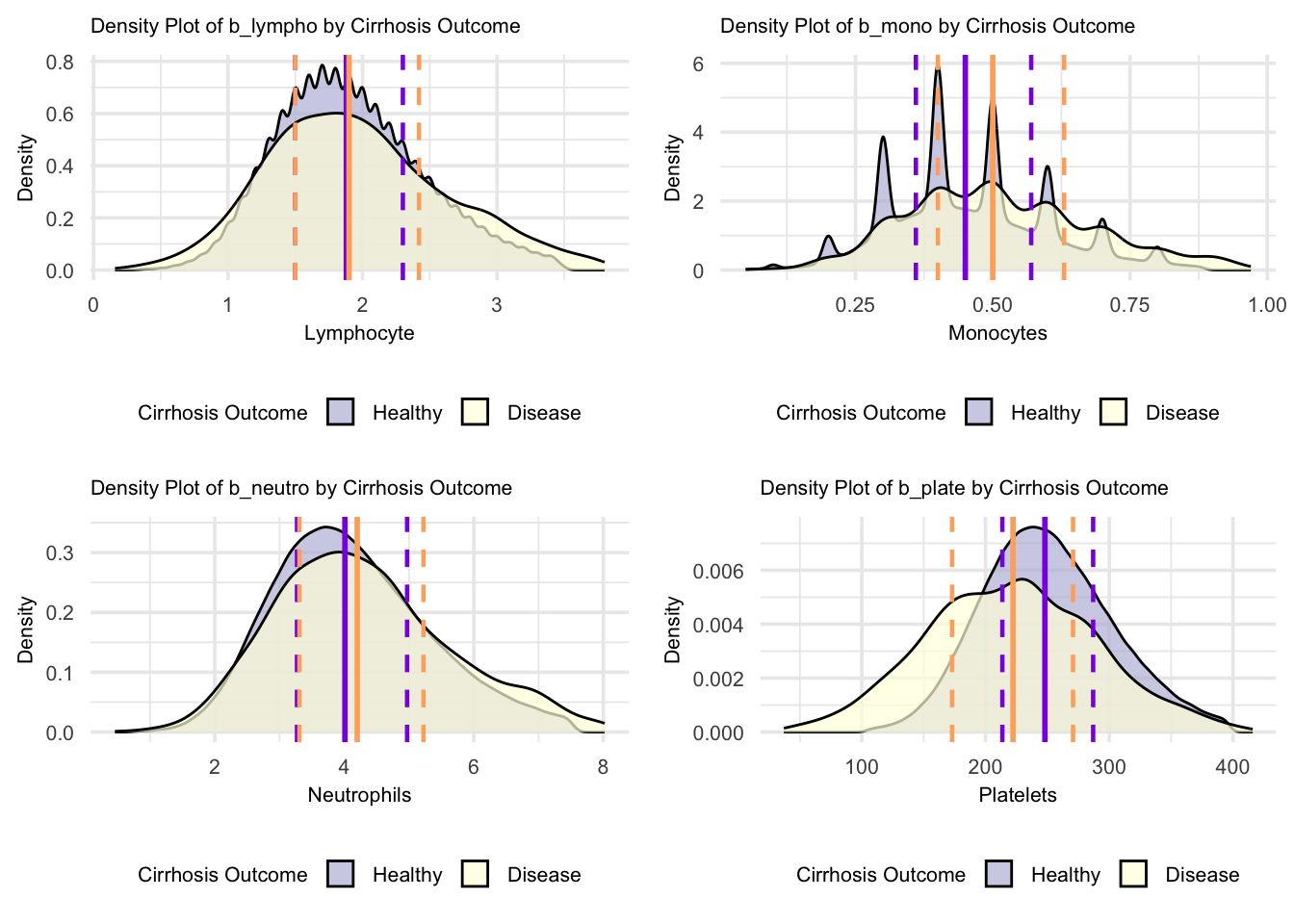
For these blood parameters, the density plots demonstrate distinctive patterns between Healthy and Cirrhosis groups. The lymphocyte distribution shows similar peak heights, while monocytes display multiple sharp peaks indicating distinct value clusters in both groups. Neutrophil distributions show broader patterns with different peak positions between groups, suggesting shifted but overlapping value ranges. The platelet distribution shows the most striking difference, with a notably higher and more concentrated peak in the healthy group compared to a lower, more dispersed distribution in cirrhosis, indicating more consistent platelet counts in health versus more variable counts in disease. Looking at the specific median lines, lymphocytes show almost identical values between healthy and cirrhosis groups, while monocytes and neutrophils demonstrate higher median values in the cirrhosis group compared to healthy individuals. In contrast, platelets show a notably higher median in the healthy group compared to cirrhosis, with the most pronounced difference among all parameters, likely reflecting the impact of portal hypertension in cirrhosis. Distribution overlaps exist between groups across all parameters, though the platelet parameter shows the clearest separation between healthy and cirrhosis populations.